3ZHC
 
 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | Descriptor: | CHLORIDE ION, FORMIC ACID, PHYTASE | | Authors: | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
8B7X
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - apo form. | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SULFATE ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Wilson, K.S, Duhme-Klair, A.K, Blagova, E.V, Bennett, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BF6
 
 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - azotochelin complex | | Descriptor: | ABC transporter, Azotochelin, FE (III) ION, ... | | Authors: | Wilson, K.S, Duhme-Klair, A.-K, Blagova, E.V, Miller, A, Booth, R, Dodson, E.J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1AX4
 
 | | TRYPTOPHANASE FROM PROTEUS VULGARIS | | Descriptor: | POTASSIUM ION, TRYPTOPHANASE | | Authors: | Isupov, M.N, Antson, A.A, Dodson, E.J, Dodson, G.G, Dementieva, I.S, Zakomirdina, L.N, Wilson, K.S, Dauter, Z, Lebedev, A.A, Harutyunyan, E.H. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of tryptophanase.
J.Mol.Biol., 276, 1998
|
|
4V20
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE, ... | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4UXH
 
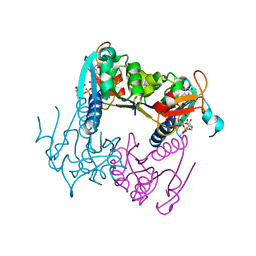 | | Leishmania major Thymidine Kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDINE KINASE, ZINC ION | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
4V6V
 
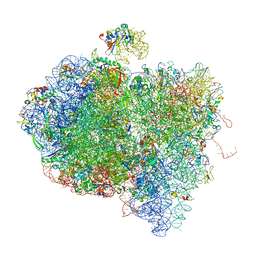 | | Tetracycline resistance protein Tet(O) bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Atkinson, G.C, Thakor, N.S, Allas, U, Lu, C, Chan, K.Y, Tenson, T, Schulten, K, Wilson, K.S, Hauryliuk, V, Frank, J. | | Deposit date: | 2013-02-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Mechanism of tetracycline resistance by ribosomal protection protein Tet(O).
Nat Commun, 4, 2013
|
|
4UXI
 
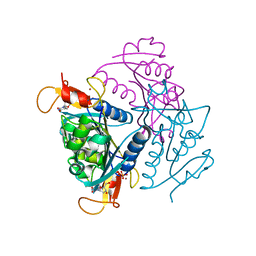 | | Leishmania major Thymidine Kinase in complex with thymidine | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
1B7V
 
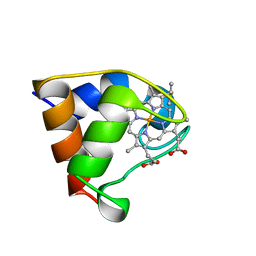 | | Structure of the C-553 cytochrome from Bacillus pasteruii to 1.7 A resolution | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C-553) | | Authors: | Gonzalez, A, Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S. | | Deposit date: | 1999-01-22 | | Release date: | 2000-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
4UBP
 
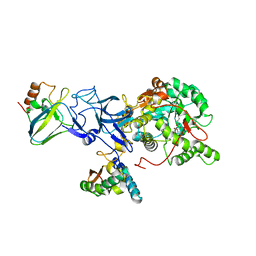 | | STRUCTURE OF BACILLUS PASTEURII UREASE INHIBITED WITH ACETOHYDROXAMIC ACID AT 1.55 A RESOLUTION | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, PROTEIN (UREASE (CHAIN A)), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1999-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The complex of Bacillus pasteurii urease with acetohydroxamate anion from X-ray data at 1.55 A resolution.
J.Biol.Inorg.Chem., 5, 2000
|
|
1BH6
 
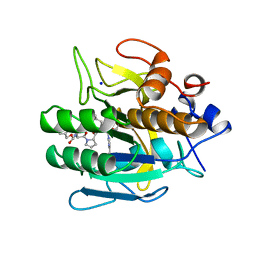 | | SUBTILISIN DY IN COMPLEX WITH THE SYNTHETIC INHIBITOR N-BENZYLOXYCARBONYL-ALA-PRO-PHE-CHLOROMETHYL KETONE | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-ALA-PRO-3-AMINO-4-PHENYL-BUTAN-2-OL, SODIUM ION, ... | | Authors: | Eschenburg, S, Genov, N, Wilson, K.S, Betzel, C. | | Deposit date: | 1998-06-15 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of subtilisin DY, a random mutant of subtilisin Carlsberg.
Eur.J.Biochem., 257, 1998
|
|
1C75
 
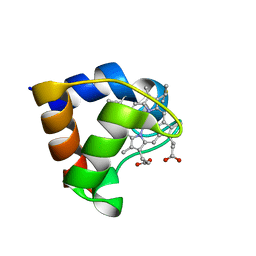 | | 0.97 A "AB INITIO" CRYSTAL STRUCTURE OF CYTOCHROME C-553 FROM BACILLUS PASTEURII | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
1CBF
 
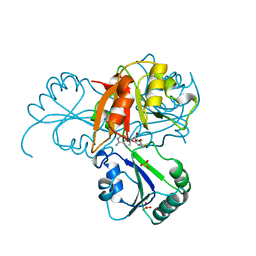 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Wilson, K.S, Warren, M.J. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
6XIA
 
 | |
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
1A48
 
 | | SAICAR SYNTHASE | | Descriptor: | PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Levdikov, V.M, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1998-02-12 | | Release date: | 1999-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
Structure, 6, 1998
|
|
5OAH
 
 | | THE PERIPLASMIC BINDING PROTEIN CEUE OF CAMPYLOBACTER JEJUNI BINDS THE IRON(III) COMPLEX OF Azotochelin | | Descriptor: | Azotochelin, Enterochelin ABC transporter substrate-binding protein, FE (III) ION | | Authors: | Raines, A.D.J, Blagova, E, Dodson, E.J, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
5OD5
 
 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | Authors: | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | Deposit date: | 2017-07-04 | | Release date: | 2018-08-01 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
1AYX
 
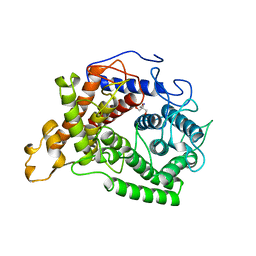 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AY7
 
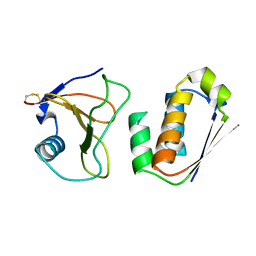 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3UBP
 
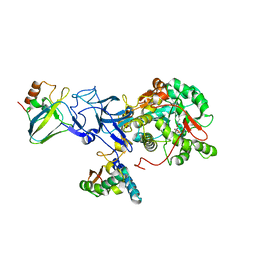 | | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | | Descriptor: | DIAMIDOPHOSPHATE, NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Miletti, S, Mangani, S, Ciurli, S. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
2WCE
 
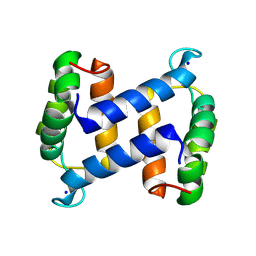 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCF
 
 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WC8
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | CITRIC ACID, PROTEIN S100-A12, SODIUM ION, ... | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCB
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | PROTEIN S100-A12, SODIUM ION, ZINC ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
