1SBX
 
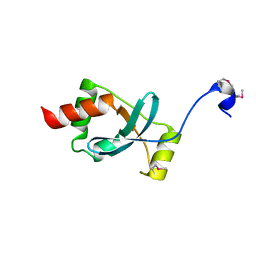 | | Crystal structure of the Dachshund-homology domain of human SKI | | Descriptor: | Ski oncogene | | Authors: | Wilson, J.J, Malakhova, M, Zhang, R, Joachimiak, A, Hegde, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Dachshund Homology Domain of human SKI
Structure, 12, 2004
|
|
2FO1
 
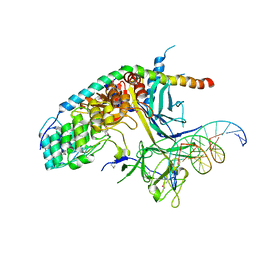 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
1NQD
 
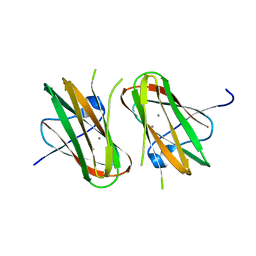 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.65 ANGSTROM RESOLUTION IN PRESENCE OF CALCIUM | | Descriptor: | CALCIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Bacterial Collagen-Binding Domain with Novel Calcium-Binding Motif Controls Domain Orientation
Embo J., 22, 2003
|
|
1NQJ
 
 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.0 ANGSTROM RESOLUTION IN ABSENCE OF CALCIUM | | Descriptor: | CHLORIDE ION, LITHIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A bacterial collagen-binding domain with novel calcium-binding motif
controls domain orientation
Embo J., 22, 2003
|
|
3BRD
 
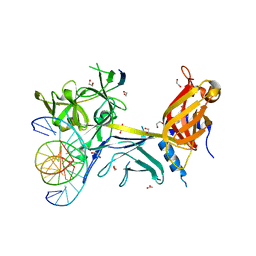 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
3BRF
 
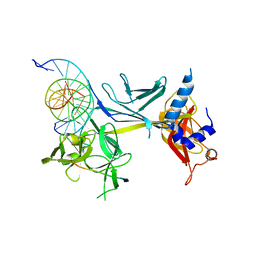 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, C2221 | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
4E37
 
 | | Crystal Structure of P. aeruginosa catalase, KatA tetramer | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | VanderWielen, B.D, Wilson, J.J, Kovall, R.A. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | KatA, the Major Catalase in Pseudomonas aeruginosa, Assists in Protecting Anaerobic Bacteria From Metabolic and Exogenous Nitric Oxide
To be Published
|
|
4FUP
 
 | |
4FUN
 
 | |
4FUM
 
 | |
4FUO
 
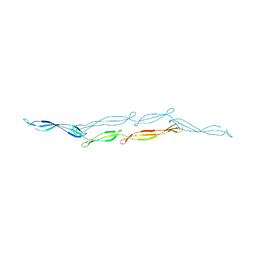 | |
4HPK
 
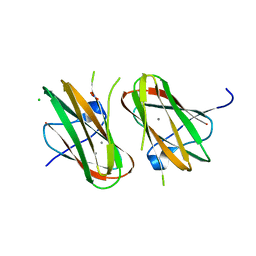 | | Crystal structure of Clostridium histolyticum colg collagenase collagen-binding domain 3B at 1.35 Angstrom resolution in presence of calcium nitrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase, ... | | Authors: | Philominathan, S.T.L, Wilson, J.J, Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
2O8O
 
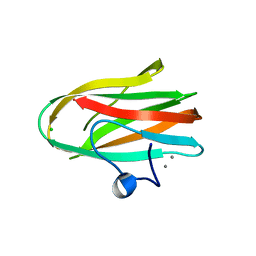 | |
6S7D
 
 | | Self-complementary duplex DNA containing an internucleoside phosphoroselenolate | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*GP*(XCI)P*CP*CP*CP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Conlon, P.F, Steinhogl, J, Vyle, J.S, Hall, J.P. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-06 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solid-phase synthesis and structural characterisation of phosphoroselenolate-modified DNA: a backbone analogue which does not impose conformational bias and facilitates SAD X-ray crystallography.
Chem Sci, 10, 2019
|
|
3BRG
 
 | | CSL (RBP-Jk) bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
3JQW
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen-binding domain 3 at 2 Angstrom resolution in presence of calcium | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Matsushita, O, Bauer, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3JQX
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen binding domain 3 at 2.2 Angstrom resolution in the presence of calcium and cadmium | | Descriptor: | CADMIUM ION, CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Bauer, R, Matsushita, O. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
