2ARL
 
 | | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding | | Descriptor: | ACETIC ACID, CHLORIDE ION, GFP-like non-fluorescent chromoprotein, ... | | Authors: | Wilmann, P.G, Battad, J, Beddoe, T, Olsen, S, Smith, S.C, Dove, S, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2005-08-19 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding
Photochem.Photobiol., 82, 2006
|
|
2G3O
 
 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
1XQM
 
 | | Variations on the GFP chromophore scaffold: A fragmented 5-membered heterocycle revealed in the 2.1A crystal structure of a non-fluorescent chromoprotein | | Descriptor: | ACETIC ACID, kindling fluorescent protein | | Authors: | Wilmann, P.G, Petersen, J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variations on the GFP chromophore: A polypeptide fragmentation within the chromophore revealed in the 2.1-A crystal structure of a nonfluorescent chromoprotein from Anemonia sulcata
J.Biol.Chem., 280, 2005
|
|
1YZW
 
 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | Descriptor: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | Authors: | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
2IE2
 
 | |
6OKJ
 
 | |
6MIS
 
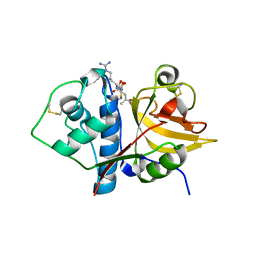 | | Native ananain in complex with E-64 | | Descriptor: | Ananain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Yongqing, T, Wilmann, P.G, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Determination of the crystal structure and substrate specificity of ananain.
Biochimie, 166, 2019
|
|
3SKO
 
 | | Crystal structure of the HLA-B8-A66-FLR, mutant A66 of the HLA B8 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SKM
 
 | | Crystal structure of the HLA-B8FLRGRAYVL, mutant G8V of the FLR peptide | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SJV
 
 | | Crystal structure of the RL42 TCR in complex with HLA-B8-FLR | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SKN
 
 | | Crystal structure of the RL42 TCR unliganded | | Descriptor: | RL42 T cell receptor, alpha chain, beta chain | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
4G8I
 
 | | Crystal Structure of HLA B2705-KK10-L6M | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, Gag protein, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G9F
 
 | | Crystal Structure of C12C TCR-HLAB2705-KK10-L6M | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G8G
 
 | | Crystal Structure of C12C TCR-HA B2705-KK10 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G9D
 
 | | Crystal Structure of HLA B2705-KK10 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4J1Y
 
 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
4KKD
 
 | | The X-ray crystal structure of Mannose-binding lectin-associated serine proteinase-3 reveals the structural basis for enzyme inactivity associated with the 3MC syndrome | | Descriptor: | IMIDAZOLE, Mannan-binding lectin serine protease 1 | | Authors: | Yongqing, T, Wilmann, P.G, Reeve, S.B, Coetzer, T.H, Smith, A.I, Whisstock, J.C, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.5991 Å) | | Cite: | The X-ray Crystal Structure of Mannose-binding Lectin-associated Serine Proteinase-3 Reveals the Structural Basis for Enzyme Inactivity Associated with the Carnevale, Mingarelli, Malpuech, and Michels (3MC) Syndrome.
J.Biol.Chem., 288, 2013
|
|
2P4M
 
 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
1UIS
 
 | | The 2.0 crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor | | Descriptor: | ACETIC ACID, CALCIUM ION, red fluorescent protein FP611 | | Authors: | Petersen, J, Wilmann, P.G, Beddoe, T, Oakley, A.J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2003-07-21 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor
J.Biol.Chem., 278, 2003
|
|
