8ENI
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
1WVN
 
 | |
3RKY
 
 | |
3RKX
 
 | |
3RKW
 
 | |
3RIR
 
 | |
7MP3
 
 | | Grb7-SH2 domain in complex with bicyclic peptide B8 | | Descriptor: | Growth factor receptor-bound protein 7, bicyclic peptide B8, {[(2R)-2,3-diamino-3-oxopropyl]sulfanyl}acetic acid | | Authors: | Colson, R, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enhancing the Bioactivity of Bicyclic Peptides Targeted to Grb7-SH2 by Restoring Cell Permeability.
Biomedicines, 10, 2022
|
|
1ZTG
 
 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
5TYI
 
 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5U1Q
 
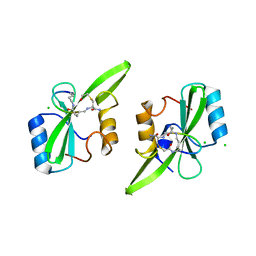 | | Grb7-SH2 with bicyclic peptide inhibitor | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 7, LYS-PHE-GLU-GLY-TYR-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
8SKK
 
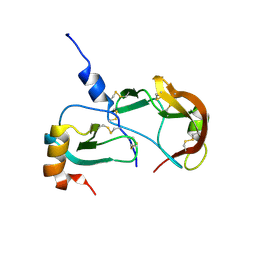 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7
To be Published
|
|
2WZM
 
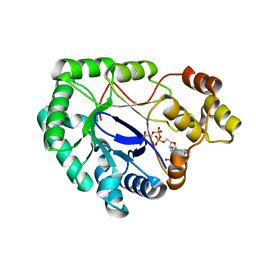 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
7S5A
 
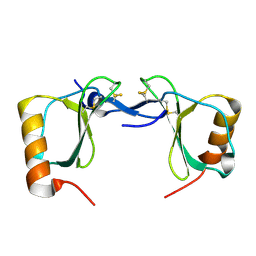 | | Crystal structure of human chemokine CCL8 | | Descriptor: | C-C motif chemokine 8 | | Authors: | Bhusal, R.P, Devkota, S.R, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-10 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6APW
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 4-[(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)methyl]benzoic acid, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
6AQQ
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[(3-fluorophenyl)methyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
2NO4
 
 | | Crystal Structure analysis of a Dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2NO5
 
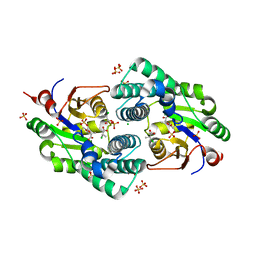 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | Descriptor: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
8FK6
 
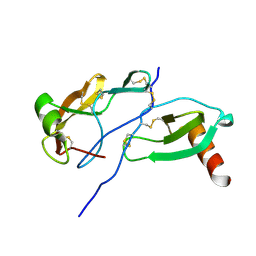 | | Crystal Structure of the Tick Evasin EVA-AAM1001(Y44A) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
2DPU
 
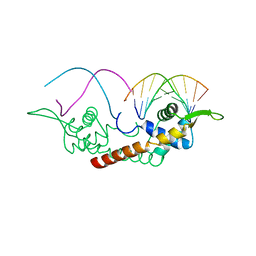 | |
2DPD
 
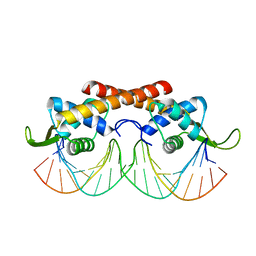 | |
2DQR
 
 | |
2EFW
 
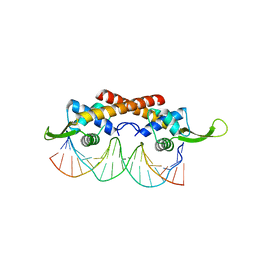 | | Crystal structure of the RTP:nRB complex from Bacillus subtilis | | Descriptor: | DNA (5'-D(*DCP*DT*DAP*DTP*DGP*DTP*DAP*DCP*DCP*DAP*DAP*DAP*DTP*DGP*DTP*DTP*DCP*DAP*DGP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DCP*DTP*DGP*DAP*DAP*DCP*DAP*DTP*DTP*DTP*DGP*DGP*DTP*DAP*DCP*DAP*DTP*DAP*DG)-3'), Replication termination protein | | Authors: | Vivian, J.P, Porter, C.J, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2007-02-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA
J.Mol.Biol., 370, 2007
|
|
3VKE
 
 | |
1T3W
 
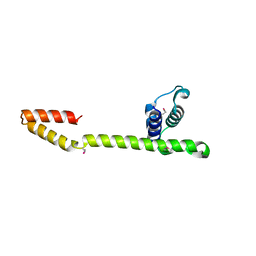 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
