1Z9B
 
 | | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tomaselli, S, Bernard, C, Spurio, R, Picone, D, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2005-04-01 | | Release date: | 2005-08-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2
Protein Sci., 14, 2005
|
|
2LYH
 
 | | Structure of Faap24 residues 141-215 | | Descriptor: | Fanconi anemia-associated protein of 24 kDa | | Authors: | Wienk, H, Slootweg, J, Kaptein, R, Boelens, R, Folkers, G.E. | | Deposit date: | 2012-09-18 | | Release date: | 2013-05-01 | | Last modified: | 2013-08-07 | | Method: | SOLUTION NMR | | Cite: | The Fanconi anemia associated protein FAAP24 uses two substrate specific binding surfaces for DNA recognition.
Nucleic Acids Res., 41, 2013
|
|
2GHF
 
 | |
2LKC
 
 | | Free B.st IF2-G2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2LKD
 
 | | IF2-G2 GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Translation initiation factor IF-2 | | Authors: | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
2MH3
 
 | | NMR structure of the basic helix-loop-helix region of the transcriptional repressor HES-1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Wienk, H, Popovic, M, Coglievina, M, Boelens, R, Pongor, S, Pintar, A. | | Deposit date: | 2013-11-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The basic helix-loop-helix region of the transcriptional repressor hairy and enhancer of split 1 is preorganized to bind DNA.
Proteins, 82, 2014
|
|
1PM6
 
 | | Solution Structure of Full-Length Excisionase (Xis) from Bacteriophage HK022 | | Descriptor: | Excisionase | | Authors: | Rogov, V.V, Luecke, C, Muresanu, L, Wienk, H, Kleinhaus, I, Werner, K, Loehr, F, Pristovsek, P, Rueterjans, H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-30 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the full-length excisionase from bacteriophage HK022.
Eur.J.Biochem., 270, 2003
|
|
1QXN
 
 | | Solution Structure of the 30 kDa Polysulfide-sulfur Transferase Homodimer from Wolinella Succinogenes | | Descriptor: | PENTASULFIDE-SULFUR, sulfide dehydrogenase | | Authors: | Lin, Y.J, Dancea, F, Loehr, F, Klimmek, O, Pfeiffer-Marek, S, Nilges, M, Wienk, H, Kroeger, A, Rueterjans, H. | | Deposit date: | 2003-09-08 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 30 kDa Polysulfide-Sulfur Transferase Homodimer from Wolinella succinogenes
Biochemistry, 43, 2004
|
|
4B5E
 
 | | Crystal Structure of an amyloid-beta binding single chain antibody PS2-8 | | Descriptor: | GLYCEROL, PS2-8, SULFATE ION | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Amyloid-Beta Binding Single Chain Antibody Ps2-8
To be Published
|
|
4B41
 
 | | Crystal structure of an amyloid-beta binding single chain antibody G7 | | Descriptor: | ANTIBODY G7, CHLORIDE ION, GLYCEROL | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Crystal Structure of an Amyloid-Beta Binding Single Chain Antibody G7
To be Published
|
|
2K16
 
 | | Solution structure of the free TAF3 PHD domain | | Descriptor: | Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2K17
 
 | | Solution structure of the TAF3 PHD domain in complex with a H3K4me3 peptide | | Descriptor: | H3K4me3 peptide, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2JVF
 
 | | Solution structure of M7, a computationally-designed artificial protein | | Descriptor: | de novo protein M7 | | Authors: | Stordeur, C, Dalluege, R, Birkenmeier, O, Wienk, H, Rudolph, R, Lange, C, Luecke, C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-08-12 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the artificial protein M7 matches the computationally designed model
Proteins, 72, 2008
|
|
2L0T
 
 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2012-03-14 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
2LRD
 
 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2NBG
 
 | | Structure of the Geobacillus stearothermophilus IF2 G3-subdomain | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Dongre, R, Folkers, G.E, Gualerzi, C.O, Boelens, R, Wienk, H. | | Deposit date: | 2016-02-11 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A model for the interaction of the G3-subdomain of Geobacillus stearothermophilus IF2 with the 30S ribosomal subunit.
Protein Sci., 25, 2016
|
|
2N01
 
 | | NMR structure of VirB9 C-terminal domain in complex with VirB7 N-terminal domain from Xanthomonas citri's T4SS | | Descriptor: | VirB7 protein, VirB9 protein | | Authors: | Oliveira, L.C, Souza, D.P, Salinas, R.K, Wienk, H, Boelens, R, Farah, S.C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-09 | | Last modified: | 2016-10-19 | | Method: | SOLUTION NMR | | Cite: | VirB7 and VirB9 Interactions Are Required for the Assembly and Antibacterial Activity of a Type IV Secretion System.
Structure, 24, 2016
|
|
2MUT
 
 | | Solution structure of the F231L mutant ERCC1-XPF dimerization region | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Faridounnia, M, Wienk, H, Kovacic, L, Folkers, G.E, Jaspers, N.G.J, Kaptein, R, Hoeijmakers, J.H.J, Boelens, R. | | Deposit date: | 2014-09-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cerebro-oculo-facio-skeletal Syndrome Point Mutation F231L in the ERCC1 DNA Repair Protein Causes Dissociation of the ERCC1-XPF Complex.
J.Biol.Chem., 290, 2015
|
|
2K4D
 
 | | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity | | Descriptor: | E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, A, De Jong, R.N, Wienk, H, Winkler, S.G, Timmers, H.T.M, Boelens, R. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity
J.Mol.Biol., 385, 2009
|
|
2KEK
 
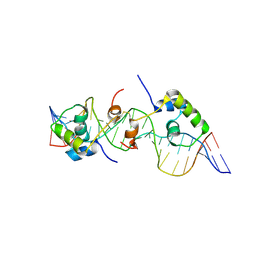 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEI
 
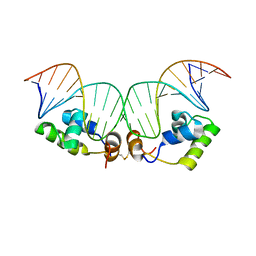 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEJ
 
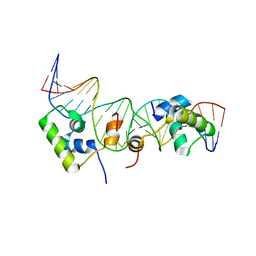 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
6YN1
 
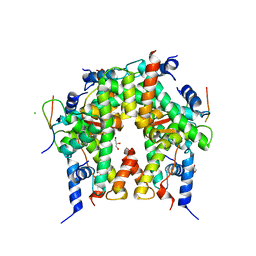 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
3KGR
 
 | |
