1CVX
 
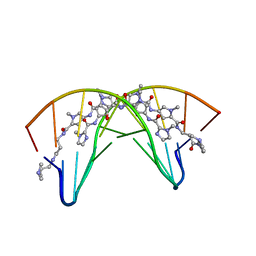 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CVY
 
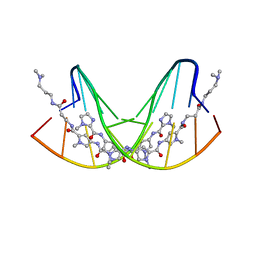 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
4R0V
 
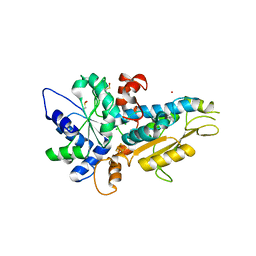 | | [FeFe]-hydrogenase Oxygen Inactivation is Initiated by the Modification and Degradation of the H cluster 2Fe Subcluster | | Descriptor: | ARSENIC, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Swanson, S.D, Ratzloff, M.W, Mulder, D.W, Artz, J.H, Ghose, S, Hoffman, A, White, S, Zadvornyy, O.A, Broderick, J.B, Bothner, B, King, P.W, Peters, J.W. | | Deposit date: | 2014-08-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster.
J.Am.Chem.Soc., 137, 2015
|
|
2TMD
 
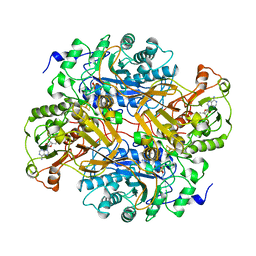 | | CORRELATION OF X-RAY DEDUCED AND EXPERIMENTAL AMINO ACID SEQUENCES OF TRIMETHYLAMINE DEHYDROGENASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Mathews, F.S, Lim, L.W, White, S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Correlation of x-ray deduced and experimental amino acid sequences of trimethylamine dehydrogenase.
J.Biol.Chem., 267, 1992
|
|
408D
 
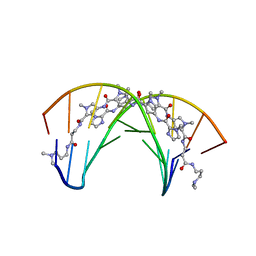 | | STRUCTURAL BASIS FOR RECOGNITION OF A-T AND T-A BASE PAIRS IN THE MINOR GROOVE OF B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*G)-3'), IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, White, S, Szewczyk, J.W, Turner, J.M, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA.
Science, 282, 1998
|
|
1ELH
 
 | |
8U11
 
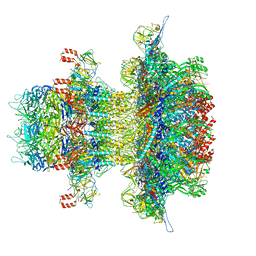 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8U10
 
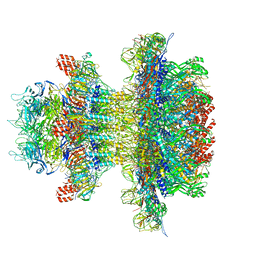 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8TVR
 
 | |
1BM9
 
 | |
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
487D
 
 | |
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
8TVU
 
 | |
8U1O
 
 | |
4AAH
 
 | | METHANOL DEHYDROGENASE FROM METHYLOPHILUS W3A1 | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE, PYRROLOQUINOLINE QUINONE | | Authors: | Mathews, F.S, Xia, Z.-X. | | Deposit date: | 1996-03-10 | | Release date: | 1996-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1.
J.Mol.Biol., 259, 1996
|
|
5W7U
 
 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 8f (SRI-29928) | | Descriptor: | 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-N-(2,3-dihydro-1H-inden-2-yl)-5-hydroxy-6-oxo-1,6-dihydropy rimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W9G
 
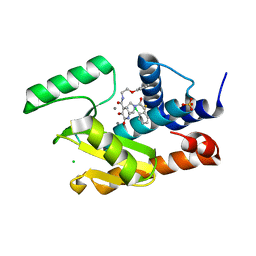 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9k (SRI-30023) | | Descriptor: | 2-[(2S)-1-{[(2-chlorophenyl)sulfanyl]acetyl}pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4 -carboxamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W73
 
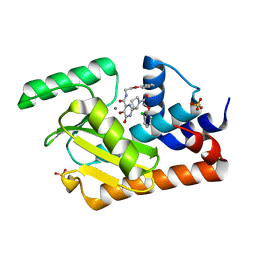 | | Crystal structure of the influenza virus PA endonuclease in complex with inhibitor 9f (SRI-29835) | | Descriptor: | 2-{(2S)-1-[(2-chlorophenoxy)acetyl]pyrrolidin-2-yl}-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
5W92
 
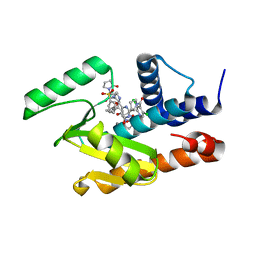 | | Crystal structure of the influenza virus PA endonuclease in complex with an inhibitor - SRI-30049 | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, 2-[(2S)-1-(3,5-dichloropyridine-4-carbonyl)pyrrolidin-2-yl]-5-hydroxy-6-oxo-N-[2-(phenylsulfonyl)ethyl]-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S. | | Deposit date: | 2017-06-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
4YWO
 
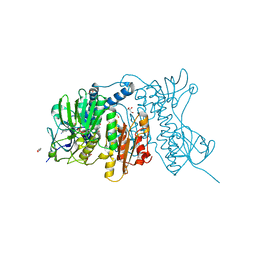 | | Mercuric reductase from Metallosphaera sedula | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase | | Authors: | Artz, J.H, Zadvornyy, O.A, White, S, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula.
Front Bioeng Biotechnol, 3, 2015
|
|
1RSS
 
 | |
1DV4
 
 | | PARTIAL STRUCTURE OF 16S RNA OF THE SMALL RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS | | Descriptor: | 16S RIBOSOMAL RNA, OCTADECATUNGSTENYL DIPHOSPHATE, RIBOSOMAL PROTEIN S5, ... | | Authors: | Tocilj, A, Schlunzen, F, Janell, D, Gluhmann, M, Hansen, H, Harms, J, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The small ribosomal subunit from Thermus thermophilus at 4.5 A resolution: pattern fittings and the identification of a functional site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1GOZ
 
 | |
