4V5E
 
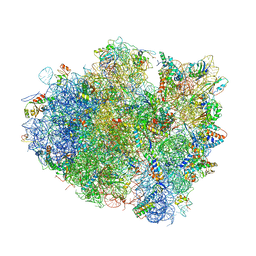 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
4V5A
 
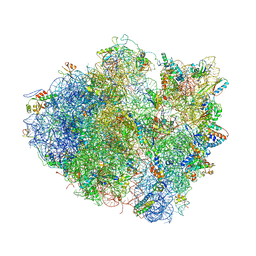 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4GZZ
 
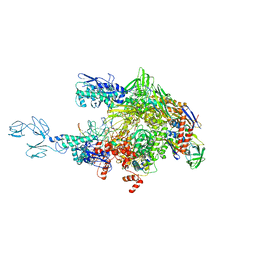 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.2927 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4GZY
 
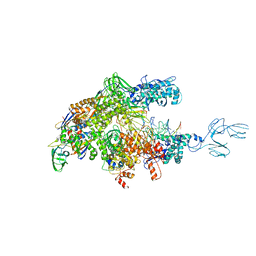 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5054 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
2UUB
 
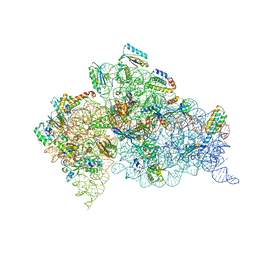 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUU-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for Expanding the Decoding Capacity of Transfer Rnas by Modification of Uridines
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UUA
 
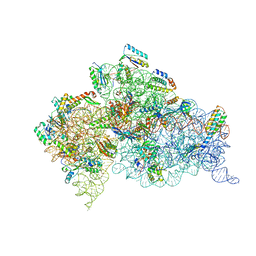 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UU9
 
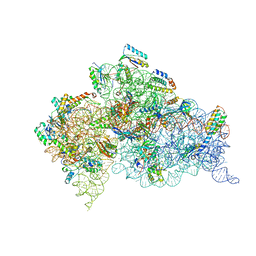 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UUC
 
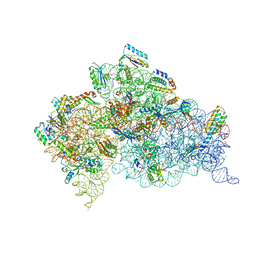 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUA-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
8AC0
 
 | |
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
2VQE
 
 | | Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U-G wobble pairing during decoding | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Kurata, S, Weixlbaumer, A, Ohtsuki, T, Shimazaki, T, Wada, T, Kirino, Y, Takai, K, Watanabe, K, Ramakrishnan, V, Suzuki, T. | | Deposit date: | 2008-03-13 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
J.Biol.Chem., 283, 2008
|
|
4V5D
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
4V5F
 
 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
6FLP
 
 | |
6FLQ
 
 | |
8ABY
 
 | |
8AC2
 
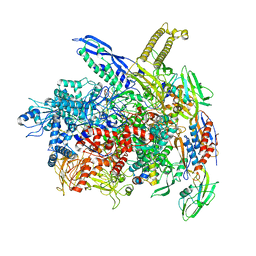 | | RNA polymerase- post-terminated, open clamp state | | Descriptor: | DNA Non-template strand, DNA Template strand, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Dey, S, Weixlbaumer, A. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into RNA-mediated transcription regulation in bacteria.
Mol.Cell, 82, 2022
|
|
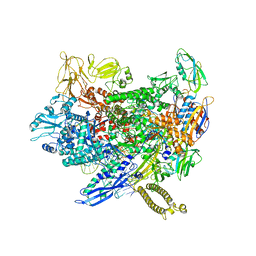
8ACP
 
 | |
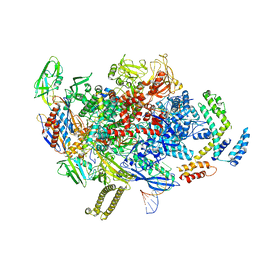
8AD1
 
 | |
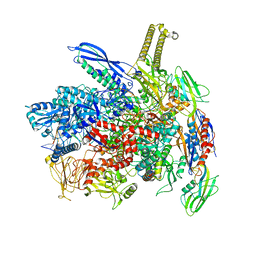
8ABZ
 
 | |
8AC1
 
 | |
7PY1
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (the consensus NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY0
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY3
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (the consensus NusA-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
