2HM8
 
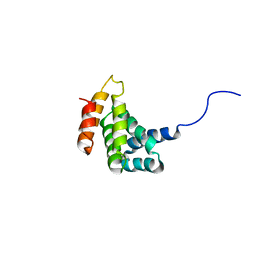 | | Solution Structure of the C-terminal MA-3 domain of Pdcd4 | | Descriptor: | Pdcd4 C-terminal MA-3 domain | | Authors: | Waters, L.C, Veverka, V, Bohm, M, Muskett, F.W, Choong, P.T, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2006-07-11 | | Release date: | 2007-02-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A
Oncogene, 26, 2007
|
|
2KZT
 
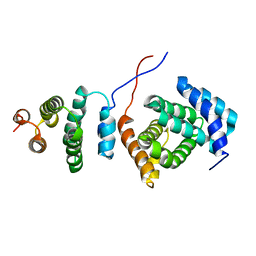 | | Structure of the Tandem MA-3 Region of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Waters, L.C, Strong, S.L, Oka, O, Muskett, F.W, Veverka, V, Banerjee, S, Schmedt, T, Henry, A.J, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2010-06-24 | | Release date: | 2011-03-16 | | Last modified: | 2011-11-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor
J.Biol.Chem., 286, 2011
|
|
5O57
 
 | | Solution Structure of the N-terminal Region of Dkk4 | | Descriptor: | Dickkopf-related protein 4 | | Authors: | Waters, L.C, Patel, S, Barkell, A.M, Muskett, F.W, Robinson, M.K, Holdsworth, G, Carr, M.D. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of Dickkopf 4 (Dkk4): New insights into Dkk evolution and regulation of Wnt signaling by Dkk and Kremen proteins.
J. Biol. Chem., 293, 2018
|
|
2M4V
 
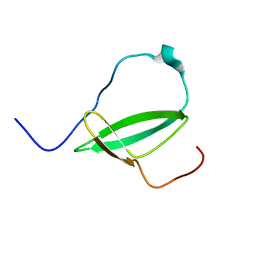 | | Mycobacterium tuberculosis RNA polymerase binding protein A (RbpA) and its interactions with sigma factors | | Descriptor: | Putative uncharacterized protein | | Authors: | Bortoluzzi, A, Muskett, F.W, Waters, L.C, Addis, P.W, Rieck, B, Munder, T, Schleier, S, Forti, F, Ghisotti, D, Carr, M.D, O'Hare, H.M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors.
J.Biol.Chem., 288, 2013
|
|
2M2D
 
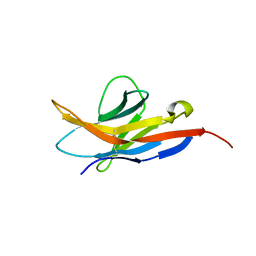 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
2KG7
 
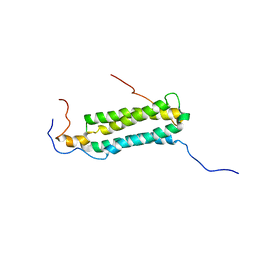 | | Structure and features of the complex formed by the tuberculosis virulence factors Rv0287 and Rv0288 | | Descriptor: | ESAT-6-like protein esxH, Uncharacterized protein esxG (PE family protein) | | Authors: | Ilghari, D, Kirsty, L.L, Waters, L.C, Veverka, V.L, Philip, R.S, Carr, M.D. | | Deposit date: | 2009-03-06 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M. tuberculosis EsxG-EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes
J.Biol.Chem., 2011
|
|
2C52
 
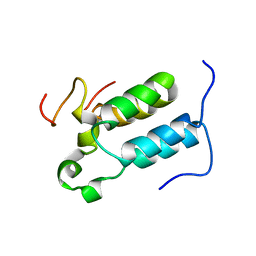 | | Structural diversity in CBP p160 complexes | | Descriptor: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | Authors: | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-15 | | Last modified: | 2018-05-02 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
8AOM
 
 | | Complex of PD-L1 with VHH1 | | Descriptor: | MAGNESIUM ION, Programmed cell death 1 ligand 1, VHH6 | | Authors: | Kang-Pettinger, T, Hall, G. | | Deposit date: | 2022-08-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Identification, binding, and structural characterization of single domain anti-PD-L1 antibodies inhibitory of immune regulatory proteins PD-1 and CD80.
J.Biol.Chem., 299, 2023
|
|
8AOK
 
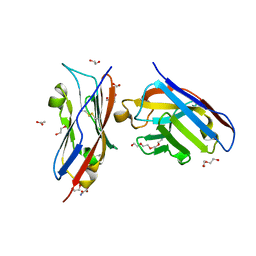 | | Complex of PD-L1 with VHH6 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kang-Pettinger, T, Hall, G. | | Deposit date: | 2022-08-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, binding, and structural characterization of single domain anti-PD-L1 antibodies inhibitory of immune regulatory proteins PD-1 and CD80.
J.Biol.Chem., 299, 2023
|
|
8CDG
 
 | | Crystal structure of human IL-17A cytokine in complex with macrocycle | | Descriptor: | (9~{S},12~{R},19~{S})-9-[[4-[[(2~{S})-2-cyclohexyl-2-(2-phenylethanoylamino)ethanoyl]amino]phenyl]methyl]-12-methyl-7,10,13,21-tetrakis(oxidanylidene)-8,11,14,20-tetrazaspiro[4.17]docosane-19-carboxylic acid, Interleukin-17A | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-01-30 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modulation of IL-17 backbone dynamics reduces receptor affinity and reveals a new inhibitory mechanism.
Chem Sci, 14, 2023
|
|
6C97
 
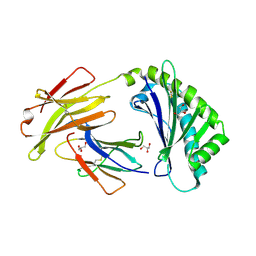 | | Crystal structure of FcRn at pH3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | Authors: | Fox III, D, Fairman, J.W. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
