4EOX
 
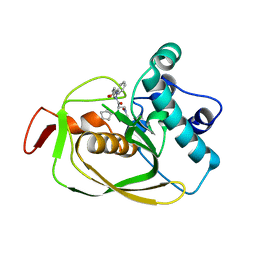 | |
5HX6
 
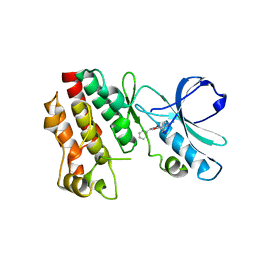 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3PYY
 
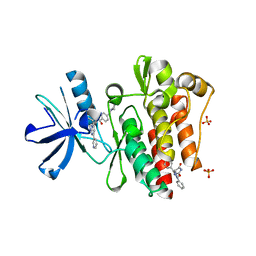 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
3SVJ
 
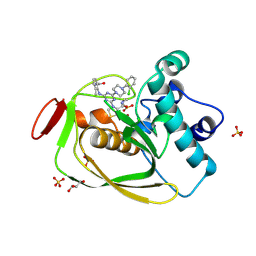 | | Strep Peptide Deformylase with a time dependent thiazolidine amide | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-methyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-12 | | Release date: | 2011-07-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
3STR
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine hydroxamic acid | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-[2-(hydroxyamino)-2-oxoethyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
3QCQ
 
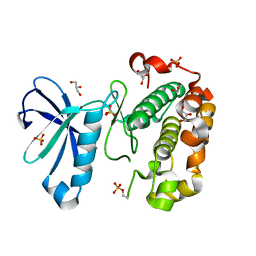 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCY
 
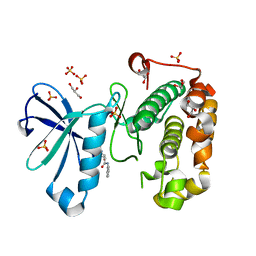 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | Descriptor: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCX
 
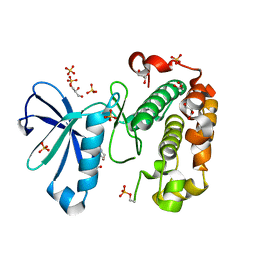 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD4
 
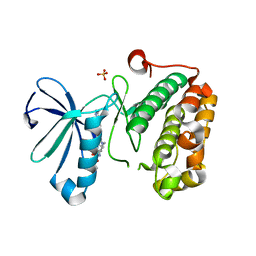 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl{(3R,5R)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-5-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, SULFATE ION, tert-butyl {(3R,5R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-5-methylpiperidin-3-yl}carbamate | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD3
 
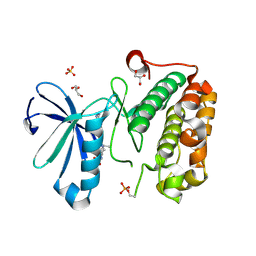 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCS
 
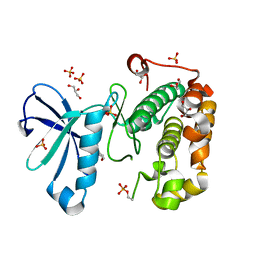 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-[2-Amino-6-(4-morpholinyl)-4-pyrimidinyl]-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-amino-6-(morpholin-4-yl)pyrimidin-4-yl]-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD0
 
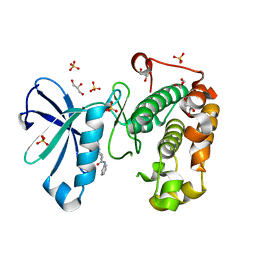 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2V84
 
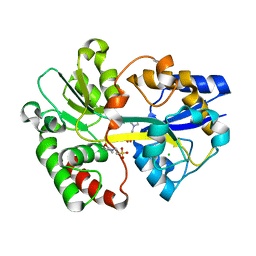 | | Crystal Structure of the Tp0655 (TpPotD) Lipoprotein of Treponema pallidum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SPERMIDINE/PUTRESCINE ABC TRANSPORTER, ... | | Authors: | Machius, M, Brautigam, C.A, Tomchick, D.R, Ward, P, Otwinowski, Z, Blevine, J.S, Deka, R.K, Norgard, M.V. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-25 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Biochemical Basis for Polyamine Binding to the Tp0655 Lipoprotein of Treponema Pallidum: Putative Role for Tp0655 (Tppotd) as a Polyamine Receptor.
J.Mol.Biol., 373, 2007
|
|
6W58
 
 | | hPGDS complexed with an aza-quinoline | | Descriptor: | 7-(azetidin-1-yl)-~{N}-[4-(2-oxidanylpropan-2-yl)cyclohexyl]-1,6-naphthyridine-3-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase | | Authors: | Elkins, P.A, Ward, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | The exploration of aza-quinolines as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors with low brain exposure.
Bioorg.Med.Chem., 28, 2020
|
|
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
6W8H
 
 | | H-PGDS complexed with inhibitor 1Y | | Descriptor: | 1,2-ETHANEDIOL, 7-cyclopropyl-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,8-naphthyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The exploration of aza-quinolines as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors with low brain exposure.
Bioorg.Med.Chem., 28, 2020
|
|
6N4E
 
 | | hPGDS complexed with a quinoline-3-carboxamide | | Descriptor: | 7-(difluoromethoxy)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]quinoline-3-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
6OW2
 
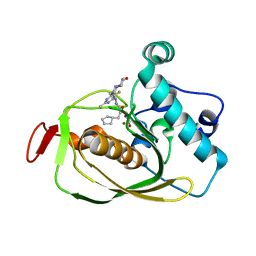 | | X-ray Structure of Polypeptide Deformylase | | Descriptor: | (2R)-2-(cyclopentylmethyl)-N'-{5-fluoro-6-[(9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]-2-methylpyrimidin-4-yl}-3-[hydroxy(hydroxymethyl)amino]propanehydrazide, NICKEL (II) ION, Peptide deformylase | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OW7
 
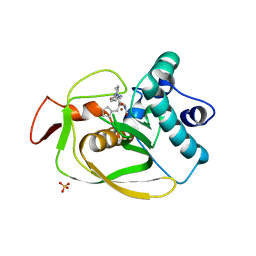 | | X-ray Structure of Polypeptide Deformylase with a Piperazic Acid | | Descriptor: | (3S)-2-{(2R)-2-(cyclopentylmethyl)-3-[formyl(hydroxy)amino]propanoyl}-N-(pyridin-2-yl)hexahydropyridazine-3-carboxamide, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3NUY
 
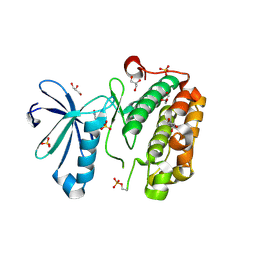 | |
3NUU
 
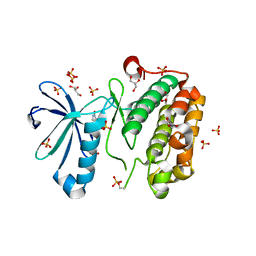 | |
3NUN
 
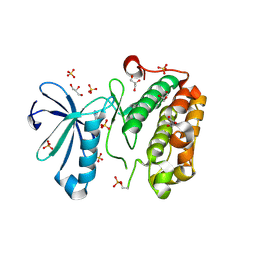 | |
3NUS
 
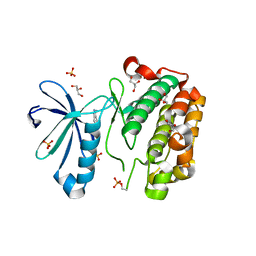 | |
4NEU
 
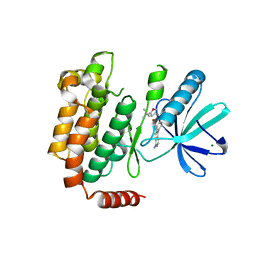 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
4CXJ
 
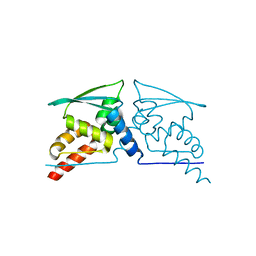 | | BTB domain of KEAP1 C151W mutant | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
