3BGG
 
 | | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Wang, X.Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Human Orotidine 5'-monophosphate Decarboxylase complexed with BMP
To be Published
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
6LQB
 
 | | A third intermediate state of L,D-transpeptidase LdtMt2-ertapenem adduct | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, D.F, Qin, Y.L. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New State I-plus Observed for the L,D-transpeptidase LdtMt2-ertapenem Adduct
Prog.Biochem.Biophys., 47, 2020
|
|
6LW2
 
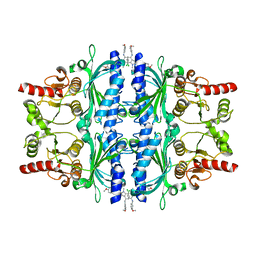 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
8HE8
 
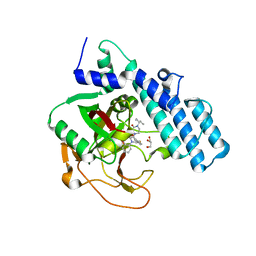 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
8HE7
 
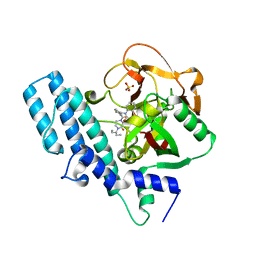 | | ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
1G7F
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177496 | | Descriptor: | 2-{4-[(2S)-2-[({[(1S)-1-CARBOXY-2-PHENYLETHYL]AMINO}CARBONYL)AMINO]-3-OXO-3-(PENTYLAMINO)PROPYL]PHENOXY}MALONIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
1G7G
 
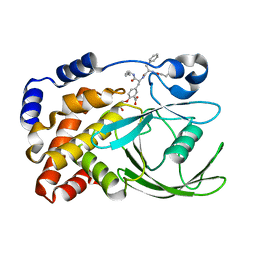 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXES WITH PNU179326 | | Descriptor: | 2-(CARBOXYMETHOXY)-5-[(2S)-2-({(2S)-2-[(3-CARBOXYPROPANOYL)AMINO] -3-PHENYLPROPANOYL}AMINO)-3-OXO-3-(PENTYLAMINO)PROPYL]BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
8WB2
 
 | | Heme-bound Arabidopsis thaliana temperature-induced lipocalin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Temperature-induced lipocalin-1 | | Authors: | Dong, C, Liu, L. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and functional studies of a plant temperature-induced lipocalin.
Biochim Biophys Acta Gen Subj, 1868, 2023
|
|
7V8P
 
 | | Crystal Structure of the MukE dimer | | Descriptor: | Chromosome partition protein MukE | | Authors: | Qian, J.W, Guo, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the chromosome partition protein MukE homodimer.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
7CKH
 
 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
8HXI
 
 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-isopropylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propan-2-ylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase L1 type 3, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HY1
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-(thiophen-2-ylmethyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(thiophen-2-ylmethyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXN
 
 | | Crystal structure of B2 Sfh-I MBL in complex with 2-amino-5-(4-(but-3-en-1-yloxy)benzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-but-3-enoxyphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Beta-lactamase, ETHANOL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXO
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-isobutylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-methylpropyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, GLYCEROL, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HY2
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-phenethylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-(2-phenylethyl)-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXW
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-heptylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-heptyl-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXE
 
 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-propylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Metallo-beta-lactamase L1 type 3, THIOCYANATE ION, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HX5
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-(4-methoxybenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-methoxyphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8HXV
 
 | | Crystal structure of B1 VIM-2 MBL in complex with 2-amino-5-hexylthiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-hexyl-1,3-thiazole-4-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
