1R0M
 
 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XPY
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-acylamino acid racemase, N~2~-ACETYL-L-GLUTAMINE | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-10 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XS2
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
3FNN
 
 | |
3GE9
 
 | |
4LXL
 
 | | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3 | | Descriptor: | H3 peptide, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | Wang, W.-C, Chu, C.-H, Chen, C.-C. | | Deposit date: | 2013-07-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3
To be Published
|
|
1FO6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE | | Descriptor: | N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE, XENON | | Authors: | Wang, W.-C, Hsu, W.-H, Chien, F.-T, Chen, C.-Y. | | Deposit date: | 2000-08-25 | | Release date: | 2001-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft.
J.Mol.Biol., 306, 2001
|
|
5WUW
 
 | |
5WUL
 
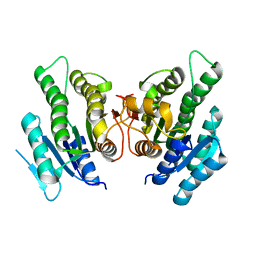 | |
4ZGW
 
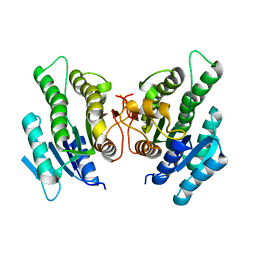 | | Short-chain dehydrogenase/reductase from Serratia marcescens BCRC 10948 | | Descriptor: | short-chain dehydrogenase/reductase | | Authors: | Wang, W.-C. | | Deposit date: | 2015-04-24 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Stereoselective synthesis of (R)-phenylephrine using recombinant
Escherichia coli cells expressing a novel short-chain
dehydrogenase/reductase gene from Serratia marcescens BCRC 10948
To Be Published
|
|
2DYU
 
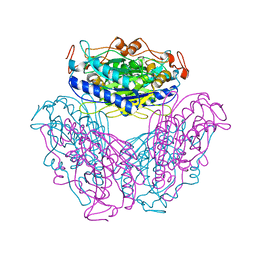 | |
2E2K
 
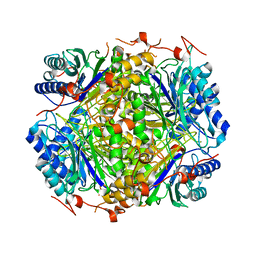 | |
2E2L
 
 | |
2DYV
 
 | |
7EQV
 
 | | Crystal structure of JMJD2A complexed with 3,4-dihydroxybenzoic acid | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | Fang, W.-K, Yang, S.-M, Wang, W.-C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Natural product myricetin is a pan-KDM4 inhibitor which with poly lactic-co-glycolic acid formulation effectively targets castration-resistant prostate cancer.
J.Biomed.Sci., 29, 2022
|
|
4YJ5
 
 | | Crystal structure of PKM2 mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
5WVA
 
 | |
