5XZ9
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, GLYCEROL | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-12 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ7
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZA
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, CITRATE ANION, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-12 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ8
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate (the ATP analog) and fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, GLYCEROL, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5XZ6
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | ATP-dependent 6-phosphofructokinase, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
4IAO
 
 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
4JYY
 
 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | Descriptor: | AZIDE ION, FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
4JZG
 
 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
4JZ2
 
 | | Crystal structure of Co ion substituted SOD2 from Clostridium difficile | | Descriptor: | COBALT (II) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Co ion substituted SOD2 from Clostridium difficile
To be Published
|
|
4M1Z
 
 | | Crystal structure of MycP1 with the N-terminal propeptide removed | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Wang, C.L, Zang, J.Y, Tian, C.L. | | Deposit date: | 2013-08-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
4GB0
 
 | |
5BOE
 
 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4OKY
 
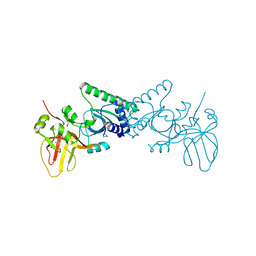 | | Crystal structure of PvuRts1I, a 5-hydroxymethylcytosine DNA restriction endonuclease | | Descriptor: | Restriction endonuclease PvuRts1 I | | Authors: | Wang, C.L, Shao, C, Zang, J.Y. | | Deposit date: | 2014-01-23 | | Release date: | 2014-09-10 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the substrate selectivity of PvuRts1I, a 5-hydroxymethylcytosine DNA restriction endonuclease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1K8U
 
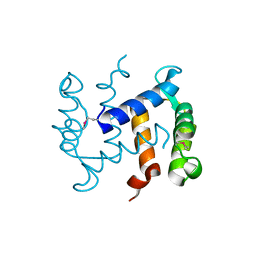 | |
1K96
 
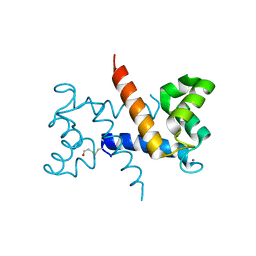 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-26 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9K
 
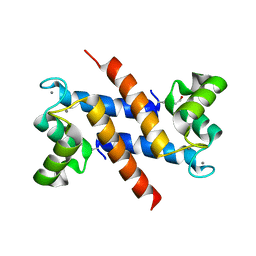 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-29 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9P
 
 | |
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2NYR
 
 | | Crystal Structure of Human Sirtuin Homolog 5 in Complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, NAD-dependent deacetylase sirtuin-5, ZINC ION | | Authors: | Min, J.R, Antoshenko, T, Allali-Hassani, A, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin.
Structure, 15, 2007
|
|
7XHO
 
 | | Structure of human inner kinetochore CCAN complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7XHN
 
 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
