4BNB
 
 | |
4BN7
 
 | |
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5I6A
 
 | | bicyclo[3.3.2]decapeptide | | Descriptor: | ALA-PHE-GLY-LYD-VAL-PHE-PRO-GLN-ALA-GLY, DIMETHYL SULFOXIDE | | Authors: | Bartoloni, M, Waltersperger, S, Bumann, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (0.813 Å) | | Cite: | Stereoselective synthesis and structure determination of a bicyclo[3.3.2]decapeptide
Arkivoc, 2014, 2014
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4I2L
 
 | | New HIV entry inhibitor MTSFT/T23 complex | | Descriptor: | GP41, inhibitor MTSFT | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-11-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.426 Å) | | Cite: | Potent antiviral activity of the novel HIV entry inhibitor MTSFT
To be Published
|
|
3VTP
 
 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
3VGX
 
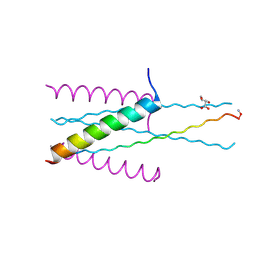 | | Structure of gp41 T21/Cp621-652 | | Descriptor: | ACETIC ACID, Envelope glycoprotein gp160, GLYCEROL | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of critical residues for viral entry and inhibition through structural Insight of HIV-1 fusion inhibitor CP621-652.
J.Biol.Chem., 287, 2012
|
|
3VH7
 
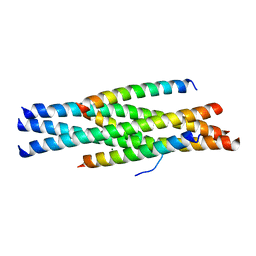 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P21 | | Descriptor: | CP32M, Envelope glycoprotein gp160, MAGNESIUM ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3VGY
 
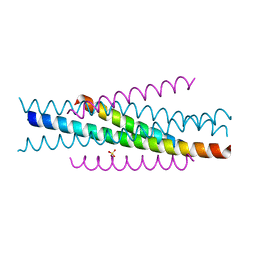 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P321 | | Descriptor: | CP32M, Envelope glycoprotein gp160, SULFATE ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3VU5
 
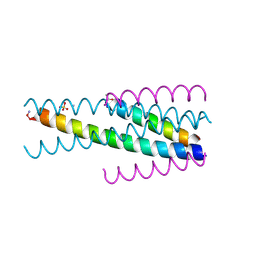 | | Short peptide HIV entry Inhibitor SC22EK | | Descriptor: | AMMONIUM ION, SC22, SULFATE ION, ... | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1
Faseb J., 27, 2013
|
|
3VWB
 
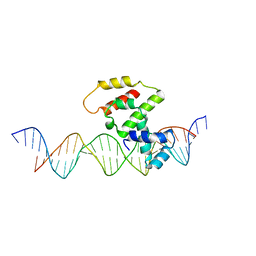 | | Crystal structure of VirB core domain (Se-Met derivative) complexed with the cis-acting site (5-BRU modifications) upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator virB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
3W19
 
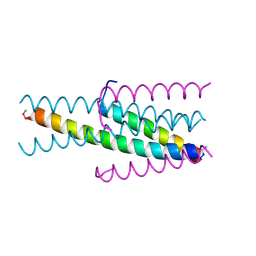 | |
3VIE
 
 | | HIV-gp41 fusion inhibitor Sifuvirtide | | Descriptor: | Envelope glycoprotein gp160, Sifuvirtide | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide
J.Biol.Chem., 287, 2012
|
|
3VTQ
 
 | | Novel HIV fusion inhibitor | | Descriptor: | Envelope glycoprotein gp160, fusion inhibitor MT-Sifuvirtide | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of a novel HIV fusion inhibitor
To be Published
|
|
3VU6
 
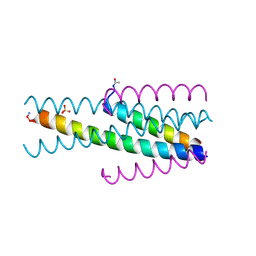 | | Short peptide HIV entry inhibitor MT-SC22EK with a M-T hook | | Descriptor: | MTSC22, SULFATE ION, Transmembrane protein gp41 | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1
Faseb J., 27, 2013
|
|
3W2A
 
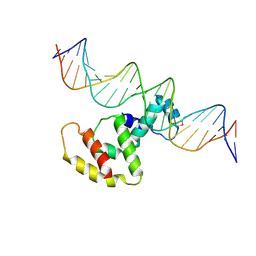 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsp promoter | | Descriptor: | DNA (31-mer), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-11-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.775 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
3W3C
 
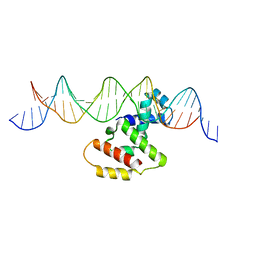 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
4BN8
 
 | |
4BN9
 
 | |
4BN6
 
 | |
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
