1HG3
 
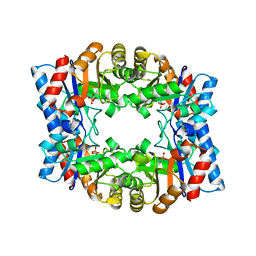 | | Crystal structure of tetrameric TIM from Pyrococcus woesei. | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Bell, G.S, Russell, R.J.M, Siebers, B, Hensel, R, Taylor, G.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tiny Tim: A Small, Tetrameric, Hyperthermostable Triosephosphate Isomerase
J.Mol.Biol., 306, 2001
|
|
1R4N
 
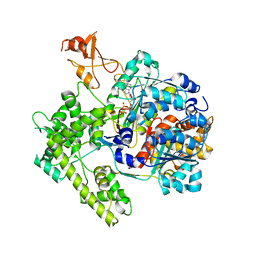 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like protein NEDD8, ZINC ION, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1R4M
 
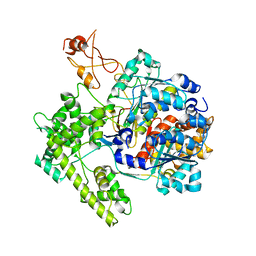 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex | | Descriptor: | Ubiquitin-like protein NEDD8, ZINC ION, amyloid beta precursor protein-binding protein 1, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1W0M
 
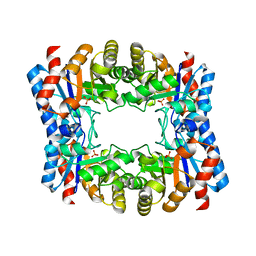 | | Triosephosphate isomerase from Thermoproteus tenax | | Descriptor: | PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Taylor, G, Lorentzen, E, Pohl, E, Lilie, H, Schramm, A, Knura, T, Stubbe, K, Tjaden, B, Hensel, R. | | Deposit date: | 2004-06-08 | | Release date: | 2004-09-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Function of a Regulated Archaeal Triosephosphate Isomerase Adapted to High Temperature
J.Mol.Biol., 342, 2004
|
|
1YOV
 
 | |
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
5OJJ
 
 | | Crystal structure of the Zn-bound ubiquitin-conjugating enzyme Ube2T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Ubiquitin-conjugating enzyme E2 T, ... | | Authors: | Morreale, F.E, Testa, A, Chaugule, V.K, Bortoluzzi, A, Ciulli, A, Walden, H. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mind the Metal: A Fragment Library-Derived Zinc Impurity Binds the E2 Ubiquitin-Conjugating Enzyme Ube2T and Induces Structural Rearrangements.
J. Med. Chem., 60, 2017
|
|
7AY0
 
 | | Crystal structure of truncated USP1-UAF1 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48, ZINC ION | | Authors: | Arkinson, C, Rennie, M.L, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AY2
 
 | | Crystal structure of truncated USP1-UAF1 reacted with ubiquitin-prg | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48, ... | | Authors: | Arkinson, C, Rennie, M.L, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AY1
 
 | | Cryo-EM structure of USP1-UAF1 bound to mono-ubiquitinated FANCD2, and FANCI | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Rennie, M.L, Arkinson, C, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5NGZ
 
 | | Ube2T in complex with fragment EM04 | | Descriptor: | 1-(1,3-benzothiazol-2-yl)methanamine, Ubiquitin-conjugating enzyme E2 T | | Authors: | Morreale, F.E, Bortoluzzi, A, Chaugule, V.K, Arkinson, C, Walden, H, Ciulli, A. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric Targeting of the Fanconi Anemia Ubiquitin-Conjugating Enzyme Ube2T by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5N2W
 
 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
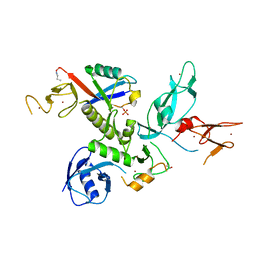 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5C23
 
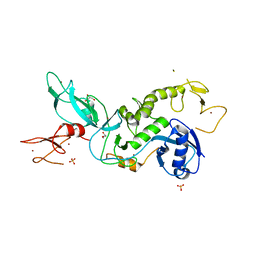 | | Parkin (S65DUblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5C1Z
 
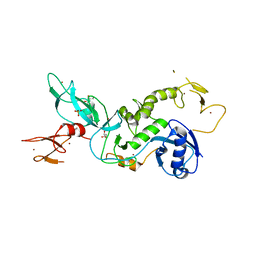 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
6R75
 
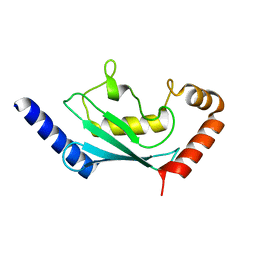 | | Crystal structure of human Ube2T E54R mutant | | Descriptor: | Ubiquitin-conjugating enzyme E2 T | | Authors: | Chaugule, V.K, Rennie, M.L, Walden, H, Arkinson, C, Kamarainen, O, Toth, R. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric mechanism for site-specific ubiquitination of FANCD2.
Nat.Chem.Biol., 16, 2020
|
|
4CCG
 
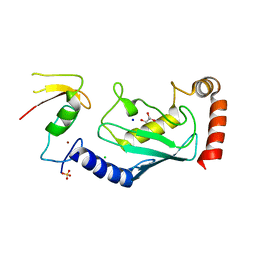 | | Structure of an E2-E3 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Hodson, C, Purkiss, A, Walden, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human Fancl Ring-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection.
Structure, 22, 2014
|
|
1W3T
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDGal, D-Glyceraldehyde and pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-LYXO-HEXONIC ACID, D-Glyceraldehyde, ... | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-19 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
7ZH3
 
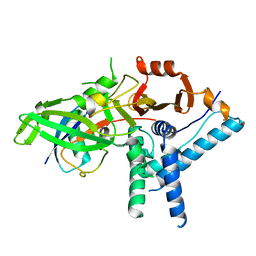 | |
7ZH4
 
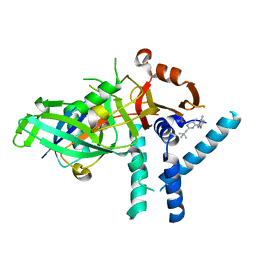 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7ZF1
 
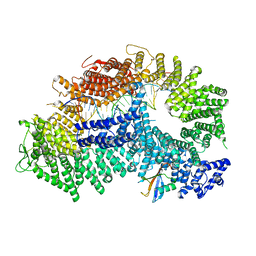 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
1W37
 
 | | 2-keto-3-deoxygluconate(KDG) aldolase of Sulfolobus solfataricus | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, SODIUM ION | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-02 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3N
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDG | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-ARABINO-HEXONIC ACID, GLYCEROL | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-17 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W3I
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with pyruvate | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, PYRUVIC ACID | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
8A9J
 
 | |
