1SPS
 
 | |
1TDE
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
1TDF
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
1SHB
 
 | |
1SPR
 
 | |
1SHA
 
 | |
7Q1V
 
 | |
8PSV
 
 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
8PTU
 
 | | 2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6Y7S
 
 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
4L0J
 
 | | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems | | Descriptor: | DNA helicase I, MAGNESIUM ION, SULFATE ION | | Authors: | Redzej, A, Ilangovan, A, Lang, S, Gruber, C.J, Topf, M, Zangger, K, Zechner, E.L, Waksman, G. | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems.
Mol.Microbiol., 89, 2013
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
1A81
 
 | |
1B7X
 
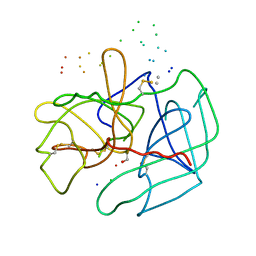 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225I MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | PROTEIN (INHIBITOR), PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-25 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4J3O
 
 | |
4KTQ
 
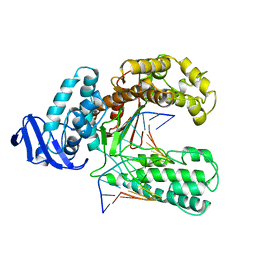 | | BINARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM T. AQUATICUS BOUND TO A PRIMER/TEMPLATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), PROTEIN (LARGE FRAGMENT OF DNA POLYMERASE I) | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-09-09 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
4AG6
 
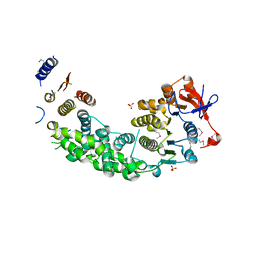 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | SULFATE ION, TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5CDW
 
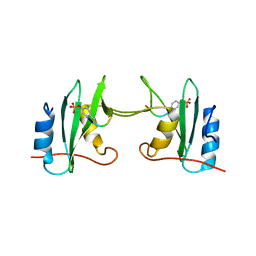 | | Crystal Structure Analysis of a mutant Grb2 SH2 domain (W121G) with a pYVNV peptide | | Descriptor: | Growth factor receptor-bound protein 2, SER-PTR-VAL-ASN-VAL-GLN | | Authors: | Papaioannou, D, Geibel, S, Kunze, M, Kay, C, Waksman, G. | | Deposit date: | 2015-07-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural and biophysical investigation of the interaction of a mutant Grb2 SH2 domain (W121G) with its cognate phosphopeptide.
Protein Sci., 25, 2016
|
|
4AG5
 
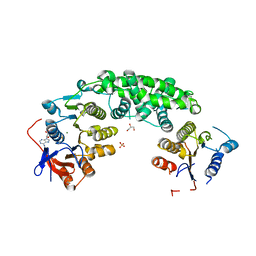 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7OIU
 
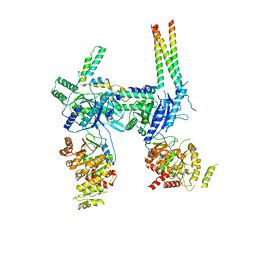 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O41
 
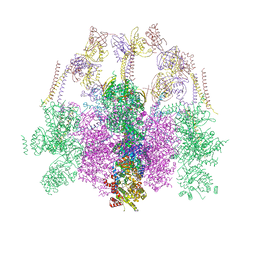 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3V
 
 | | Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwI protein, TrwJ protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O42
 
 | | TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwK protein,Protein hcp1 | | Authors: | Vadakkepat, A.K, Mace, K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
