6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYG
 
 | | Crystal Structure of 5-(trifluoromethoxy)indoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-(trifluoromethyloxy)-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6TVN
 
 | | Crystal Structure of 5-bromoindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase | | Descriptor: | 5-bromanyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
7OWG
 
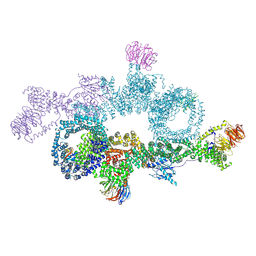 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
6RIQ
 
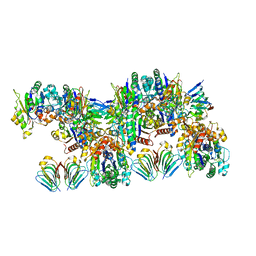 | | MinCD filament from Pseudomonas aeruginosa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MinC, ... | | Authors: | Szewczak-Harris, A, Wagstaff, J, Lowe, J. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the MinCD copolymeric filament from Pseudomonas aeruginosa at 3.1 angstrom resolution.
Febs Lett., 593, 2019
|
|
6TT2
 
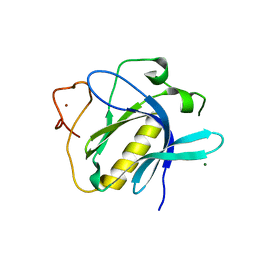 | |
6TSE
 
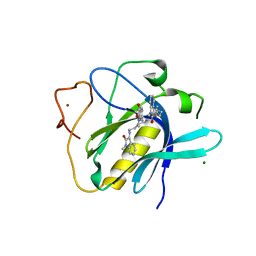 | | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1-methylindole-2,3-dione, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TUH
 
 | | The PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
7QUD
 
 | | D. melanogaster alpha/beta tubulin heterodimer in the GTP form | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tubulin alpha-1 chain, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUC
 
 | | D. melanogaster alpha/beta tubulin heterodimer in the GDP form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUQ
 
 | | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii) | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin beta, Tubulin domain-containing protein | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
7QUP
 
 | | D. melanogaster 13-protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
4WZP
 
 | | Ser65 phosphorylated ubiquitin, major conformation | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Wauer, T, Wagstaff, J, Freund, S.M.V, Komander, D. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis.
Embo J., 34, 2015
|
|
7OWC
 
 | |
7OWD
 
 | |
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
5LJN
 
 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
6TL1
 
 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
5QUE
 
 | |
5QUF
 
 | |
5QUB
 
 | |
5QUC
 
 | |
5QUH
 
 | |
5QUD
 
 | |
