5NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6TRG
 
 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
6NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
359D
 
 | |
5D65
 
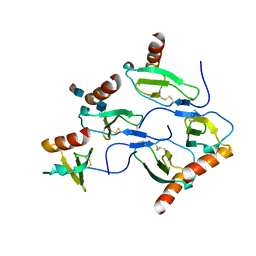 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6XOS
 
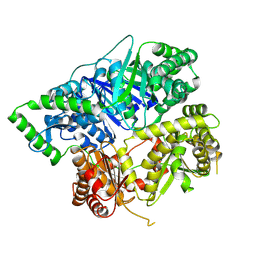 | |
6XOU
 
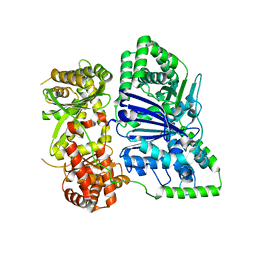 | |
6XOV
 
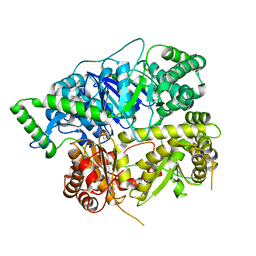 | |
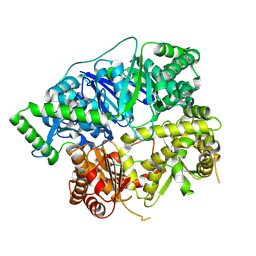
6XOW
 
 | |
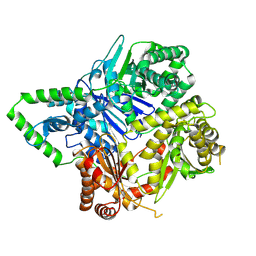
6XOT
 
 | |
5DNF
 
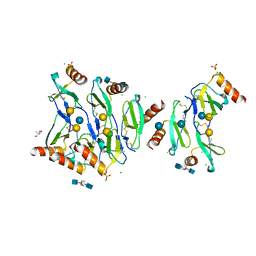 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GZ4
 
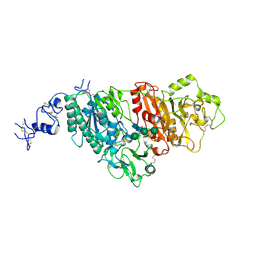 | |
5GZ5
 
 | |
3NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
5HQ8
 
 | |
5HI7
 
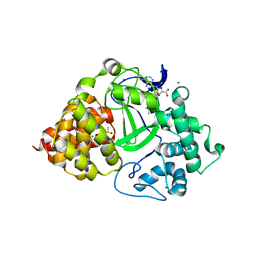 | | Co-crystal structure of human SMYD3 with an aza-SAH compound | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][3-(dimethylamino)propyl]amino}-5'-deoxyadenosine, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Bonnette, W.G. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets.
Structure, 24, 2016
|
|
5I7J
 
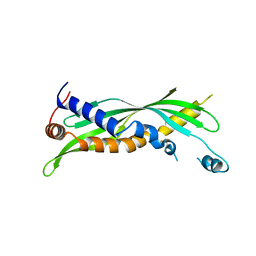 | |
5I7K
 
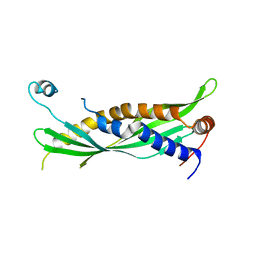 | | Crystal Structure of Human SPLUNC1 Dolphin Mutant D1 (G58A, S61A, G62E, G63D, G66D, I67T) | | Descriptor: | BPI fold-containing family A member 1 | | Authors: | Walton, W.G, Redinbo, M.R. | | Deposit date: | 2016-02-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1.
Biochemistry, 55, 2016
|
|
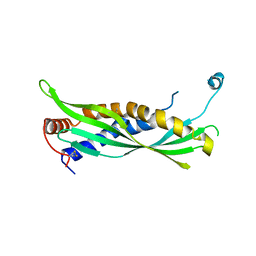
5I7L
 
 | |
4CSB
 
 | | Structure of the Virulence-Associated Protein VapD from the intracellular pathogen Rhodococcus equi. | | Descriptor: | VIRULENCE ASSOCIATED PROTEIN VAPD, octyl beta-D-glucopyranoside | | Authors: | Whittingham, J.L, Blagova, E.V, Finn, C.E, Luo, H, Miranda-CasoLuengo, R, Turkenburg, J.P, Leech, A.P, Walton, P.H, Meijers, W.G, Wilkinson, A.J. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Virulence-Associated Protein Vapd from the Intracellular Pathogen Rhodococcus Equi.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6NCZ
 
 | |
6NCY
 
 | | Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans | | Descriptor: | Beta-glucuronidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selecting a Single Stereocenter: The Molecular Nuances That Differentiate beta-Hexuronidases in the Human Gut Microbiome.
Biochemistry, 58, 2019
|
|
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
