6XFK
 
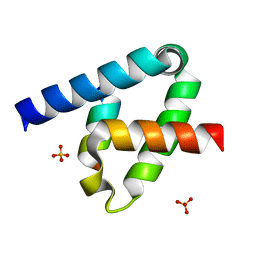 | | Crystal structure of the type III secretion system pilotin-secretin complex InvH-InvG | | Descriptor: | SULFATE ION, Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFJ
 
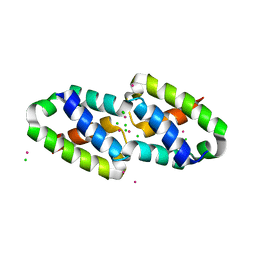 | | Crystal structure of the type III secretion pilotin InvH | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XFL
 
 | | Structural characterization of the type III secretion system pilotin-secretin complex InvH-InvG by NMR spectroscopy | | Descriptor: | Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6DUZ
 
 | | Structure of the periplasmic domains of PrgH and PrgK from the assembled Salmonella type III secretion injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
1YJ7
 
 | | Crystal structure of enteropathogenic E.coli (EPEC) type III secretion system protein EscJ | | Descriptor: | GLYCEROL, PHOSPHATE ION, escJ | | Authors: | Yip, C.K, Kimbrough, T.G, Felise, H.B, Vuckovic, M, Thomas, N.A, Pfuetzner, R.A, Frey, E.A, Finlay, B.B, Miller, S.I, Strynadka, N.C.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the molecular platform for type III secretion system assembly.
Nature, 435, 2005
|
|
5TCP
 
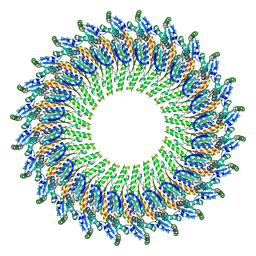 | | Near-atomic resolution cryo-EM structure of the periplasmic domains of PrgH and PrgK | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
8EXQ
 
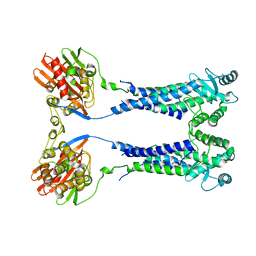 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXP
 
 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXR
 
 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
4G2S
 
 | |
2OBL
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ACETATE ION, CALCIUM ION, EscN, ... | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OBM
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, EscN | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5TCQ
 
 | | Near-atomic resolution cryo-EM structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG | | Descriptor: | Protein InvG | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
5TCR
 
 | | Atomic model of the Salmonella SPI-1 type III secretion injectisome basal body proteins InvG, PrgH, and PrgK | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
2X49
 
 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
2X4A
 
 | |
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | Descriptor: | Protein PrgI | | Authors: | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV3
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV6
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG (residues 176-end) in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
3T07
 
 | | Crystal structure of S. aureus Pyruvate Kinase in complex with a naturally occurring bis-indole alkaloid | | Descriptor: | (3S,5R)-3,5-bis(6-bromo-1H-indol-3-yl)piperazin-2-one, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities.
J.Biol.Chem., 286, 2011
|
|
3T05
 
 | |
6NJP
 
 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
