1NSO
 
 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
2NPU
 
 | | The solution structure of the rapamycin-binding domain of mTOR (FRB) | | Descriptor: | FKBP12-rapamycin complex-associated protein | | Authors: | Veverka, V, Crabbe, T, Bird, I, Lennie, G, Muskett, F.W, Taylor, R.J, Carr, M.D. | | Deposit date: | 2006-10-30 | | Release date: | 2007-09-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR.
Oncogene, 27, 2008
|
|
2K8P
 
 | | Characterisation of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation | | Descriptor: | Sclerostin | | Authors: | Veverka, V, Henry, A.J, Slocombe, P.M, Ventom, A, Mulloy, B, Muskett, F.W, Muzylak, M, Greenslade, K, Moore, A, Zhang, L, Gong, J, Qian, X, Paszty, C, Taylor, R.J, Robinson, M.K, Carr, M.D. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Structural Features and Interactions of Sclerostin: Molecular insight into a key regulator of Wnt-mediated bone formation
J.Biol.Chem., 284, 2009
|
|
2L3Y
 
 | | Solution structure of mouse IL-6 | | Descriptor: | Interleukin-6 | | Authors: | Veverka, V, Redpath, N.T, Carrington, B, Muskett, F.W, Taylor, R.J, Henry, A.J, Carr, M.D. | | Deposit date: | 2010-09-25 | | Release date: | 2011-09-28 | | Last modified: | 2013-01-02 | | Method: | SOLUTION NMR | | Cite: | Conservation of functional sites on interleukin-6 and implications for evolution of signaling complex assembly and therapeutic intervention.
J.Biol.Chem., 287, 2012
|
|
5YI9
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 56-91) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Cell division cycle-associated 7-like protein | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMO
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 1-32) complex | | Descriptor: | PC4 and SFRS1-interacting protein,LEDGF/p75 IBD-JPO2 M1 | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMP
 
 | | Solution structure of the LEDGF/p75 IBD - POGZ (aa 1370-1404) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Pogo transposable element with ZNF domain | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMQ
 
 | | Solution structure of the LEDGF/p75 IBD - MLL1 (aa 111-160) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Histone-lysine N-methyltransferase 2A | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMR
 
 | |
5K57
 
 | | HDD domain from human Ddi2 | | Descriptor: | Protein DDI1 homolog 2 | | Authors: | Veverka, V. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Human DNA-Damage-Inducible 2 Protein Is Structurally and Functionally Distinct from Its Yeast Ortholog.
Sci Rep, 6, 2016
|
|
6ZV0
 
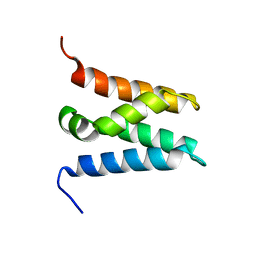 | | TFIIS N-terminal domain (TND) from human LEDGF/p75 | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV4
 
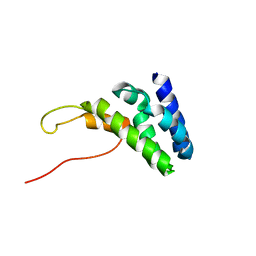 | | Human TFIIS N-terminal domain in complex with IWS1 | | Descriptor: | Transcription elongation factor A protein 1,Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV3
 
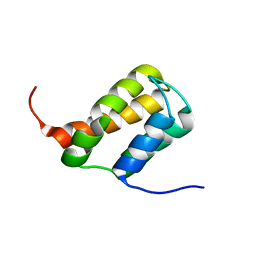 | | TFIIS N-terminal domain (TND) from human MED26 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV1
 
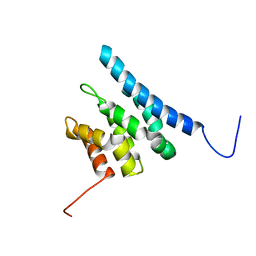 | | TFIIS N-terminal domain (TND) from human IWS1 | | Descriptor: | Protein IWS1 homolog | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZUY
 
 | | Human TFIIS N-terminal domain (TND) | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZUZ
 
 | |
6ZV2
 
 | | TFIIS N-terminal domain (TND) from human PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Veverka, V. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
2N73
 
 | | Solution structure of the ACBD3:PI4KB complex | | Descriptor: | Golgi resident protein GCP60, Phosphatidylinositol 4-kinase beta | | Authors: | Veverka, V, Hexnerova, R. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
2N72
 
 | | Solution structure of the Q domain from ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | Veverka, V, Hexnerova, R. | | Deposit date: | 2015-09-02 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
6T3I
 
 | | Solution structure of the HRP2 IBD | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Veverka, V. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Unlike Its Paralog LEDGF/p75, HRP-2 Is Dispensable for MLL-R Leukemogenesis but Important for Leukemic Cell Survival.
Cells, 10, 2021
|
|
6YI3
 
 | |
7ACS
 
 | |
7ACT
 
 | |
2N2X
 
 | |
2N2W
 
 | |
