2V6N
 
 | | Crystal structures of the SARS-coronavirus main proteinase inactivated by benzotriazole compounds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(DIMETHYLAMINO)BENZOIC ACID, REPLICASE POLYPROTEIN 1AB, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-07-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
2VJ1
 
 | | A Structural View of the Inactivation of the SARS-Coronavirus Main Proteinase by Benzotriazole Esters | | Descriptor: | 4-(DIMETHYLAMINO)BENZOIC ACID, BENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
6Z2H
 
 | |
6ZNW
 
 | |
8P8Q
 
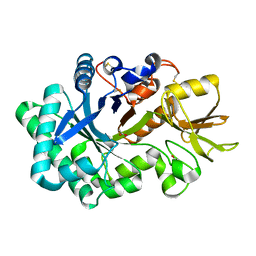 | | Recombinant Ym1 crystal structure | | Descriptor: | ACETATE ION, Chitinase-like protein 3, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Aegerter, H, Smole, U, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8T
 
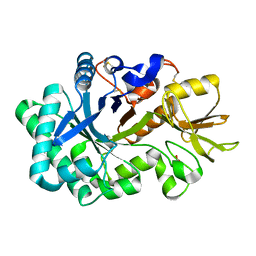 | | Ex vivo Ym2 crystal structure | | Descriptor: | Chitinase-like protein 4, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8S
 
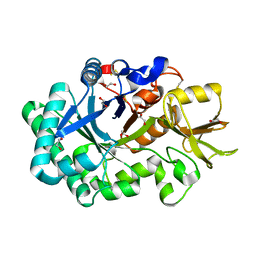 | | Recombinant Ym2 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 4 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, A, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8P8R
 
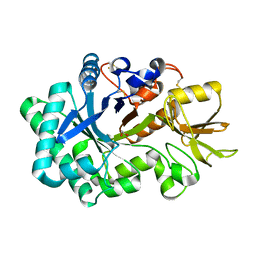 | | Ex vivo Ym1 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-like protein 3 | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
2BX3
 
 | |
2BX4
 
 | |
7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
6QRN
 
 | | Galectin-10 complexed with ribose | | Descriptor: | Galectin-10, beta-D-ribopyranose | | Authors: | Verstraete, K, Verschueren, K.H.G. | | Deposit date: | 2019-02-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
1ODS
 
 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODT
 
 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
6QCL
 
 | |
6QFB
 
 | |
1AL3
 
 | |
6HXL
 
 | |
6HXO
 
 | |
6HXJ
 
 | |
6HXP
 
 | |
