4C0R
 
 | | Molecular and structural basis of glutathione import in Gram-positive bacteria via GshT and the cystine ABC importer TcyBC of Streptococcus mutans. | | Descriptor: | CADMIUM ION, CHLORIDE ION, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Vergauwen, B, Verstraete, K, Senadheera, D.B, Dansercoer, A, Cvitkovitch, D.G, Guedon, E, Savvides, S.N. | | Deposit date: | 2013-08-07 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Molecular and Structural Basis of Glutathione Import in Gram-Positive Bacteria Via Gsht and the Cystine Abc Importer Tcybc of Streptococcus Mutans.
Mol.Microbiol., 89, 2013
|
|
3M8U
 
 | | Crystal structure of glutathione-binding protein A (GbpA) from Haemophilus parasuis SH0165 in complex with glutathione disulfide (GSSG) | | Descriptor: | Heme-binding protein A, MALONATE ION, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Vergauwen, B, Elegheert, J, Dansercoer, A, Devreese, B, Savvides, S.N. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TPA
 
 | |
2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
2R9Z
 
 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
3LN7
 
 | |
3LN6
 
 | |
4PU8
 
 | | Shewanella oneidensis Toxin Antitoxin System Antitoxin Protein HipB Resolution 2.35 | | Descriptor: | Toxin-antitoxin system antidote transcriptional repressor Xre family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4PU3
 
 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P212121) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4PU5
 
 | | Shewanella oneidensis Toxin Antitoxin System Toxin Protein HipA Bound with AMPPNP and Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4PU4
 
 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P21) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.786 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4PU7
 
 | | Shewanella oneidensis Toxin Antitoxin System Antitoxin Protein HipB Resolution 1.85 | | Descriptor: | Toxin-antitoxin system antidote transcriptional repressor Xre family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
3UCE
 
 | | Crystal structure of a small-chain dehydrogenase in complex with NADPH | | Descriptor: | Dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Buysschaert, G, Verstraete, K, Savvides, S, Vergauwen, B. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference.
Febs J., 280, 2013
|
|
3UCF
 
 | |
2BE7
 
 | | Crystal structure of the unliganded (T-state) aspartate transcarbamoylase of the psychrophilic bacterium Moritella profunda | | Descriptor: | Aspartate Carbamoyltransferase Catalytic Chain, Aspartate Carbamoyltransferase Regulatory Chain, SULFATE ION, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda.
J.Mol.Biol., 365, 2007
|
|
3QS9
 
 | |
3QS7
 
 | |
5D28
 
 | |
5D22
 
 | |
8C7L
 
 | |
3UF2
 
 | |
3UF5
 
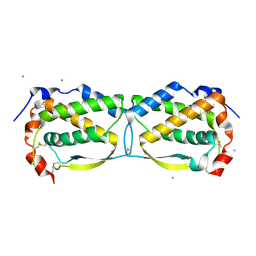 | | Crystal structure of the mouse Colony-Stimulating Factor 1 (mCSF-1) cytokine | | Descriptor: | CALCIUM ION, Macrophage colony-stimulating factor 1 | | Authors: | Elegheert, J, Bracke, N, Bekaert, A, Savvides, S.N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-08-22 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEZ
 
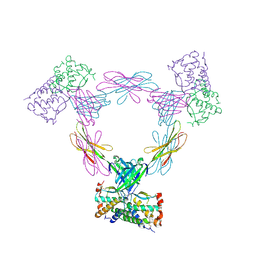 | |
4ADQ
 
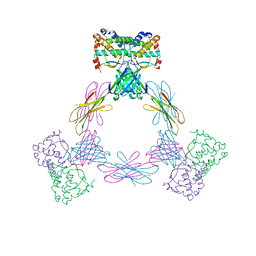 | | CRYSTAL STRUCTURE OF THE MOUSE COLONY-STIMULATING FACTOR 1 (MCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2012-01-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ADF
 
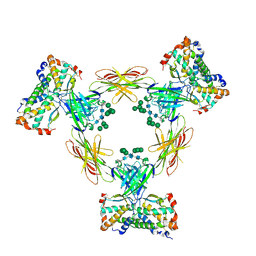 | | CRYSTAL STRUCTURE OF THE HUMAN COLONY-STIMULATING FACTOR 1 (hCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2011-12-23 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
