4R9U
 
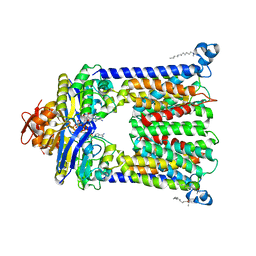 | | Structure of vitamin B12 transporter BtuCD in a nucleotide-bound outward facing state | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Korkhov, V.M, Mireku, S.A, Veprintsev, D.B, Locher, K.P. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3HSY
 
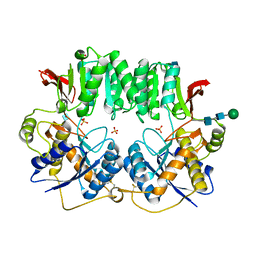 | | High resolution structure of a dimeric GluR2 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, SULFATE ION, ... | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
3O2J
 
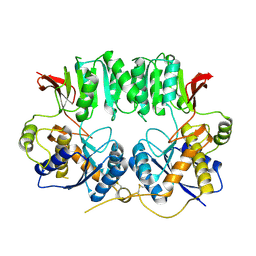 | | Structure of the GluA2 NTD-dimer interface mutant, N54A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2010-07-22 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
1UPT
 
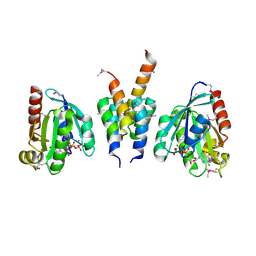 | | Structure of a complex of the golgin-245 GRIP domain with Arl1 | | Descriptor: | ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 1, GOLGI AUTOANTIGEN, GOLGIN SUBFAMILY A MEMBER 4, ... | | Authors: | Panic, B, Perisic, O, Veprintsev, D.B, Williams, R.L, Munro, S. | | Deposit date: | 2003-10-12 | | Release date: | 2003-10-30 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Arl1-Dependent Targeting of Homodimeric Grip Domains to the Golgi Apparatus
Mol.Cell, 12, 2003
|
|
2J9V
 
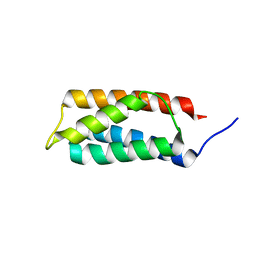 | | 2 Angstrom X-ray structure of the yeast ESCRT-I Vps28 C-terminus | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 28 | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2J9W
 
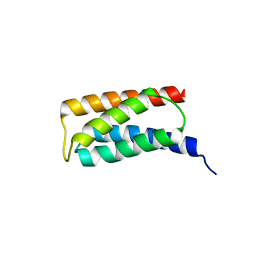 | | Structural insight into the ESCRT-I-II link and its role in MVB trafficking | | Descriptor: | VPS28-PROV PROTEIN | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2J9U
 
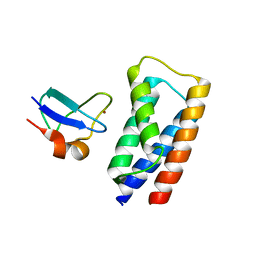 | | 2 Angstrom X-ray structure of the yeast ESCRT-I Vps28 C-terminus in complex with the NZF-N domain from ESCRT-II | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 28, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 36, ZINC ION | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2CAZ
 
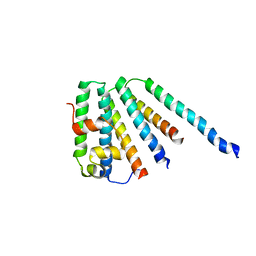 | | ESCRT-I core | | Descriptor: | PROTEIN SRN2, SUPPRESSOR PROTEIN STP22 OF TEMPERATURE-SENSITIVE ALPHA-FACTOR RECEPTOR AND ARGININE PERMEASE, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN VPS28 | | Authors: | Gill, D.J, Teo, H, Sun, J, Perisic, O, Veprintsev, D.B, Vallis, Y, Emr, S.D, Williams, R.L. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
4I1Q
 
 | | Crystal Structure of hBRAP1 N-BAR domain | | Descriptor: | Bridging integrator 2 | | Authors: | Sanchez-Barrena, M.J. | | Deposit date: | 2012-11-21 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Bin2 Is a Membrane Sculpting N-BAR Protein That Influences Leucocyte Podosomes, Motility and Phagocytosis
Plos One, 7, 2012
|
|
3SL9
 
 | | X-ray structure of Beta catenin in complex with Bcl9 | | Descriptor: | 1,2-ETHANEDIOL, B-cell CLL/lymphoma 9 protein, Catenin beta-1, ... | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
3SLA
 
 | | X-ray structure of first four repeats of human beta-catenin | | Descriptor: | Catenin beta-1, GLYCEROL, SODIUM ION | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
1OA8
 
 | |
3N6V
 
 | |
2VUK
 
 | |
2WQJ
 
 | |
2WQI
 
 | |
2WUJ
 
 | | DivIVA N-terminal domain | | Descriptor: | SEPTUM SITE-DETERMINING PROTEIN DIVIVA | | Authors: | Oliva, M.A, Leonard, T.A, Lowe, J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Features Critical for Membrane Binding Revealed by Diviva Crystal Structure.
Embo J., 29, 2010
|
|
2WTT
 
 | |
2WUK
 
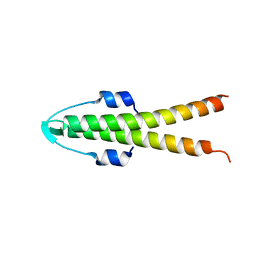 | |
2YBG
 
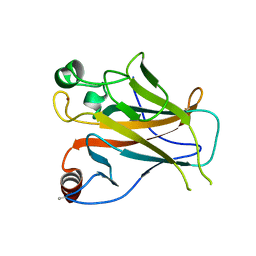 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1W2D
 
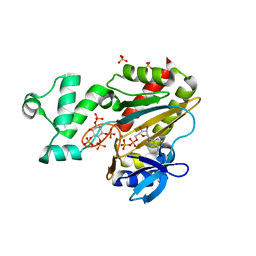 | | Human Inositol (1,4,5)-trisphosphate 3-kinase complexed with Mn2+/ADP/Ins(1,3,4,5)P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W2F
 
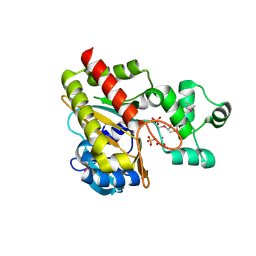 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W2C
 
 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
2BIN
 
 | |
2BIP
 
 | |
