7PQP
 
 | | tau-microtubule structural ensemble based on CryoEM data | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Isoform Tau-F of Microtubule-associated protein tau, ... | | Authors: | Brotzakis, Z.F, Vendruscolo, M. | | Deposit date: | 2021-09-18 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A Structural Ensemble of a Tau-Microtubule Complex Reveals Regulatory Tau Phosphorylation and Acetylation Mechanisms.
Acs Cent.Sci., 7, 2021
|
|
7PQC
 
 | | tau-microtubule structural ensemble based on CryoEM data | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Isoform Tau-F of Microtubule-associated protein tau, ... | | Authors: | Brotzakis, Z.F, Vendruscolo, M. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A Structural Ensemble of a Tau-Microtubule Complex Reveals Regulatory Tau Phosphorylation and Acetylation Mechanisms.
Acs Cent.Sci., 7, 2021
|
|
6O2T
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2S
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2Q
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2NR2
 
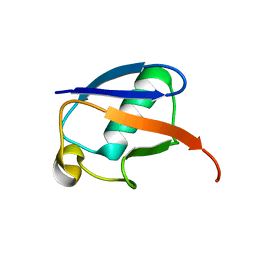 | | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native states ensembles of proteins | | Descriptor: | Ubiquitin | | Authors: | Richter, B, Gsponer, J, Varnai, P, Salvatella, X, Vendruscolo, M. | | Deposit date: | 2006-11-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native state ensembles of proteins
J.Biomol.Nmr, 37, 2007
|
|
5J0M
 
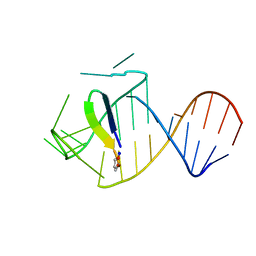 | | Ground state sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29-mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-28 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J1O
 
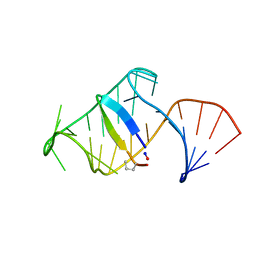 | | Excited state (Bound-like) sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29mer) of the HIV-1 TAR element, Cyclic peptide mimetic of Tat | | Authors: | Borkar, A.N, Bardaro, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J2W
 
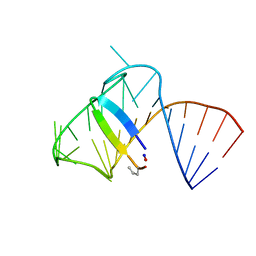 | | Intermediate state lying on the pathway of release of Tat from HIV-1 TAR. | | Descriptor: | Apical region (29mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1XQQ
 
 | | Simultaneous determination of protein structure and dynamics | | Descriptor: | ubiquitin | | Authors: | Lindorff-Larsen, K, Best, R.B, DePristo, M.A, Vendruscolo, M, Dobson, C.M. | | Deposit date: | 2004-10-13 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Simultaneous determination of protein structure and dynamics
Nature, 433, 2005
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6G4A
 
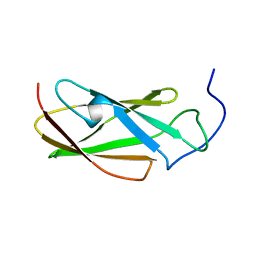 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
2K0P
 
 | |
2LPF
 
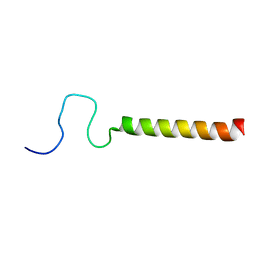 | | R state structure of monomeric phospholamban (C36A, C41F, C46A) | | Descriptor: | Cardiac phospholamban | | Authors: | De Simone, A, Montalvao, R.W, Gustavsson, M, Shi, L, Veglia, G, Vendruscolo, M. | | Deposit date: | 2012-02-11 | | Release date: | 2013-02-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures of the Excited States of Phospholamban and Shifts in Their Populations upon Phosphorylation.
Biochemistry, 52, 2013
|
|
2L2P
 
 | | Folding Intermediate of the Fyn SH3 A39V/N53P/V55L from NMR Relaxation Dispersion Experiments | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2010-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2MOR
 
 | |
2K0E
 
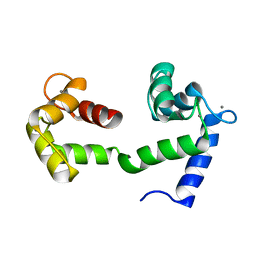 | | A Coupled Equilibrium Shift Mechanism in Calmodulin-Mediated Signal Transduction | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Gsponer, J, Christodoulou, J, Cavalli, A, Bui, J.M, Richter, B, Dobson, C.M, Vendruscolo, M. | | Deposit date: | 2008-02-02 | | Release date: | 2008-06-10 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Structure, 16, 2008
|
|
2K0F
 
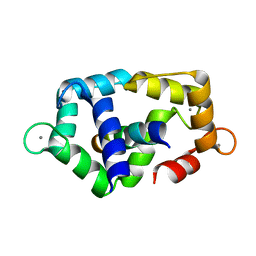 | | Calmodulin complexed with calmodulin-binding peptide from smooth muscle myosin light chain kinase | | Descriptor: | 19-mer peptide from Myosin light chain kinase, CALCIUM ION, calmodulin | | Authors: | Gsponer, J, Christodoulou, J, Cavalli, A, Bui, J.M, Richter, B, Dobson, C.M, Vendruscolo, M. | | Deposit date: | 2008-02-02 | | Release date: | 2008-06-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Structure, 16, 2008
|
|
2K5X
 
 | |
2LJ5
 
 | |
2LP5
 
 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2MEZ
 
 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2014-08-20 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
2M5N
 
 | | Atomic-resolution structure of a cross-beta protofilament | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-{beta} amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M5M
 
 | | Atomic-resolution structure of a triplet cross-beta amyloid fibril | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (12.2 Å), SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-beta amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
