5OJ8
 
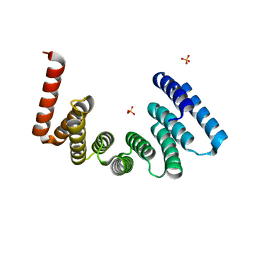 | | Crystal structure of the KLC1-TPR domain ([A1-B5] fragment) | | Descriptor: | Kinesin light chain 1, PHOSPHATE ION | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
5OJF
 
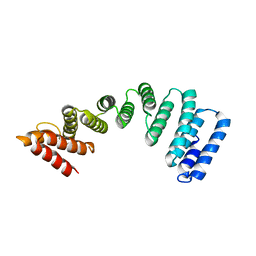 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
4UUZ
 
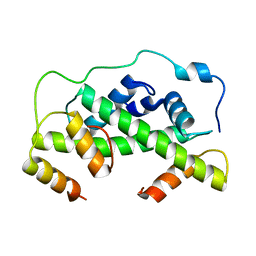 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
6T66
 
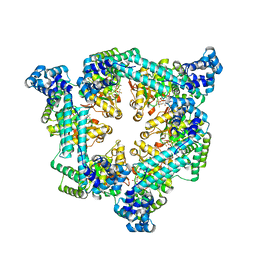 | | Crystal structure of the Vibrio cholerae replicative helicase (DnaB) with GDP-AlF4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Legrand, P, Quevillon-Cheruel, S, Li de la Sierra-Gallay, I, Walbott, H. | | Deposit date: | 2019-10-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res., 49, 2021
|
|
6HWP
 
 | |
4P3Z
 
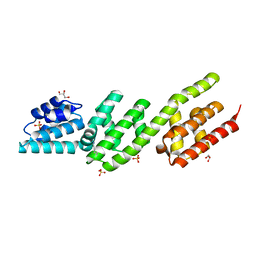 | | Chlamydia pneumoniae CopN (D29 construct) | | Descriptor: | CopN, GLYCEROL, SULFATE ION | | Authors: | Nawrotek, A, Guimaraes, B.G, Knossow, M, Gigant, B. | | Deposit date: | 2014-03-10 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
J.Biol.Chem., 289, 2014
|
|
4P40
 
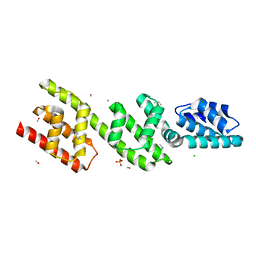 | | Chlamydia pneumoniae CopN | | Descriptor: | CHLORIDE ION, CopN, DIMETHYLAMINE, ... | | Authors: | Nawrotek, A, Guimaraes, B.G, Knossow, M, Gigant, B. | | Deposit date: | 2014-03-10 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae.
J.Biol.Chem., 289, 2014
|
|
4ZA6
 
 | |
5OC6
 
 | |
5OC5
 
 | |
5OC4
 
 | | Crystal structure of human tRNA-dihydrouridine(20) synthase dsRBD R361A-R362A mutant | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bou-nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2017-06-29 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular basis for transfer RNA recognition by the double-stranded RNA-binding domain of human dihydrouridine synthase 2.
Nucleic Acids Res., 47, 2019
|
|
7A3Z
 
 | | OSM-3 kinesin motor domain complexed with Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Osmotic avoidance abnormal protein 3 | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
7A40
 
 | | Nucleotide-free OSM-3 kinesin motor domain | | Descriptor: | GLYCEROL, Osmotic avoidance abnormal protein 3, SULFATE ION | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
7A5E
 
 | | OSM-3 kinesin motor domain complexed with Mg.AMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Osmotic avoidance abnormal protein 3, ... | | Authors: | Varela, F.P, Menetrey, J, Gigant, B. | | Deposit date: | 2020-08-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural snapshots of the kinesin-2 OSM-3 along its nucleotide cycle: implications for the ATP hydrolysis mechanism.
Febs Open Bio, 11, 2021
|
|
6FT5
 
 | | Structure of A3_A3, an artificial bi-domain protein based on two identical alphaRep A3 domains | | Descriptor: | GLYCEROL, SULFATE ION, alphaRep A3_A3 | | Authors: | Li de la Sierra-Gallay, I, Leger, C, Di Meo, T. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 9, 2019
|
|
6FSQ
 
 | |
6F7T
 
 | |
6GX7
 
 | | Tubulin-CopN-alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-26 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVN
 
 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVM
 
 | | Tubulin:F3II DARPin complex | | Descriptor: | F3II DARPIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V, Cantos Fernandes, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FI9
 
 | |
8BJI
 
 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR1
 
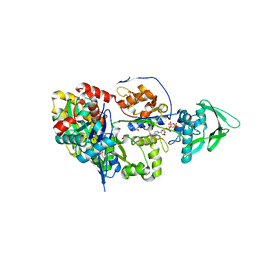 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR0
 
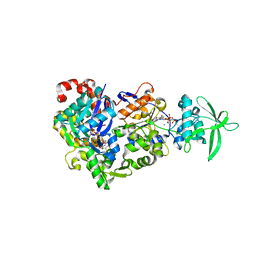 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin (residue Q3455 to L3863) in complex with 3'deoxyCTP and two manganese cations bound to Latrunculin-B-ADP-Mn-actin | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Texeira-Nuns, M, Retailleau, P, Renault, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BO1
 
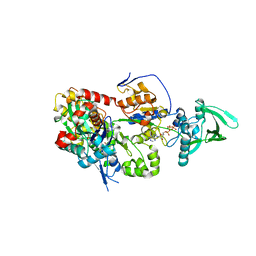 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
