2BAX
 
 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | Deposit date: | 2005-10-15 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1C8Q
 
 | |
2IRP
 
 | | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Putative aldolase class 2 protein aq_1979 | | Authors: | Jeyakanthan, J, Gayathri, D, Yogavel, M, Velmurugan, D, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5
To be Published
|
|
5GIB
 
 | | Succinic acid bound trypsin crystallized as dimer | | Descriptor: | CALCIUM ION, Cationic trypsin, SUCCINIC ACID | | Authors: | Kutumbarao, N.H.V, Manohar, R, KarthiK, L, Malathy, P, Gunasekaran, K, Velmurugan, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Trypsin bound with Succinic acid
To Be Published
|
|
1GH4
 
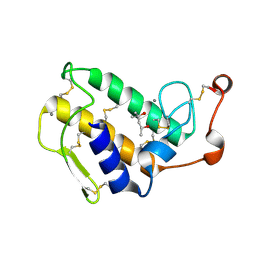 | | Structure of the triple mutant (K56M, K120M, K121M) of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2000-11-09 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Observation of additional calcium ion in the crystal structure of the triple mutant K56,120,121M of bovine pancreatic phospholipase A2.
J.Mol.Biol., 324, 2002
|
|
5JYI
 
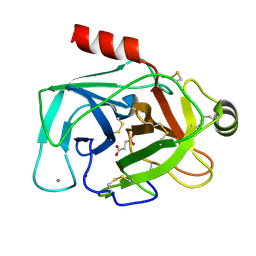 | | Trypsin bound with succinic acid at 1.9A | | Descriptor: | CALCIUM ION, Cationic trypsin, SODIUM ION, ... | | Authors: | Manohar, R, Kutumbarao, N.H.V, KarthiK, L, Malathy, P, Velmurugan, D, Gunasekaran, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Trypsin bound with succinic acid at 1.9A
To Be Published
|
|
1VL9
 
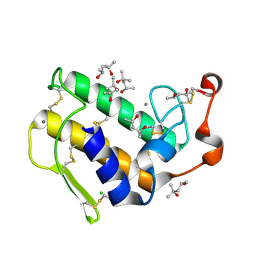 | | Atomic resolution (0.97A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Gayathri, D, Poi, M.-J, Tsai, M.-D, Dauter, M, Dauter, Z. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4U02
 
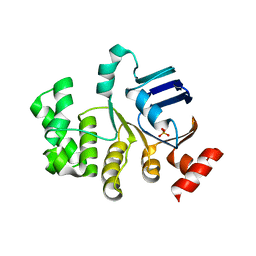 | | Crystal structure of apo-TTHA1159 | | Descriptor: | Amino acid ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
4U00
 
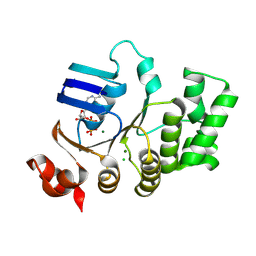 | | Crystal structure of TTHA1159 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Amino acid ABC transporter, ATP-binding protein, ... | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
4H1P
 
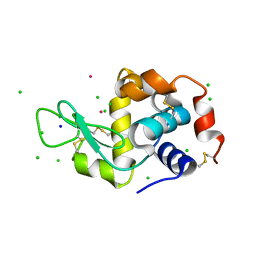 | |
1VKQ
 
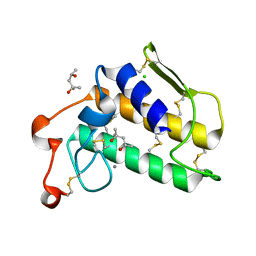 | | A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Yamane, T, Dauter, M, Dauter, Z. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1O2E
 
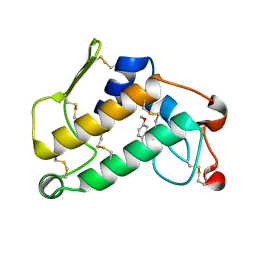 | | Structure of the triple mutant (K53,56,120M) + Anisic acid complex of phospholipase A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the free and anisic acid bound triple mutant of phospholipase A2.
J.Mol.Biol., 333, 2003
|
|
2OM6
 
 | | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable phosphoserine phosphatase, ... | | Authors: | Jeyakanthan, J, Vaijayanthimala, S, Gayathri, D, Velmurugan, D, Baba, S, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hypothetical Protein (Probable Phosphoserine Phosph (PH0253) from Pyrococcus Horikoshii OT3
To be Published
|
|
2PCR
 
 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
2BCH
 
 | | A possible of Second calcium ion in interfacial binding: Atomic and Medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Yogavel, M, Velmurugan, D, Poi, M.J, Dauter, Z, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BD1
 
 | | A possible role of the second calcium ion in interfacial binding: Atomic and medium resolution crystal structures of the quadruple mutant of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Phospholipase A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Suggestive evidence for the involvement of the second calcium and surface loop in interfacial binding: monoclinic and trigonal crystal structures of a quadruple mutant of phospholipase A(2).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2PT5
 
 | | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Shikimate kinase | | Authors: | Jeyakanthan, J, Nithya, N, Shimada, A, Velmurugan, D, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5
To be Published
|
|
2PKP
 
 | | Crystal structure of 3-isopropylmalate dehydratase (leuD)from Methhanocaldococcus Jannaschii DSM2661 (MJ1271) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Homoaconitase small subunit, ZINC ION | | Authors: | Jeyakanthan, J, Gayathri, D.R, Velmurugan, D, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-18 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity determinants of the methanogen homoaconitase enzyme: structure and function of the small subunit
Biochemistry, 49, 2010
|
|
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
1O3W
 
 | |
3P5W
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P5U
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P5X
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
