4OO9
 
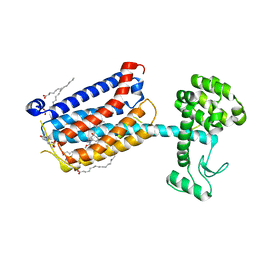 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | Authors: | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
2PZS
 
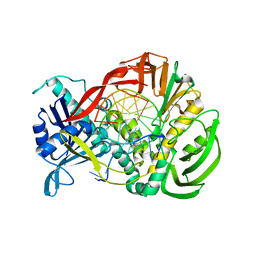 | | Phi29 DNA polymerase complexed with primer-template DNA (post-translocation binary complex) | | Descriptor: | 5'-d(CTAACACGTAAGCAGTC)-3', 5'-d(GACTGCTTAC)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PY5
 
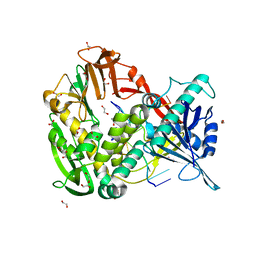 | | Phi29 DNA polymerase complexed with single-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(GGACTTT)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYL
 
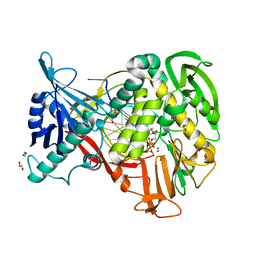 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(CTGACGAATGTACA)-3', 5'-d(GACTGCTTAC(2DA))-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PYJ
 
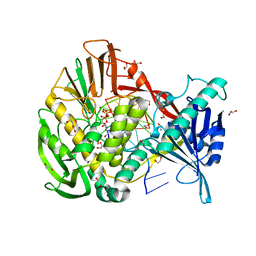 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-d(ACACGTAAGCAGTC)-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2C1C
 
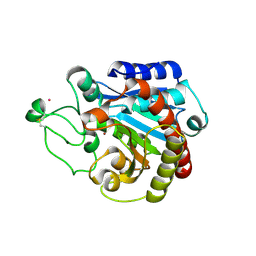 | | Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors | | Descriptor: | CARBOXYPEPTIDASE B, YTTRIUM ION, ZINC ION | | Authors: | Bayes, A, Comellas-Bigler, M, Rodriguez de la Vega, M, Maskos, K, Bode, W, Aviles, F.X, Jongsma, M.A, Beekwilder, J, Vendrell, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Resistance of an Insect Carboxypeptidase to Plant Protease Inhibitors.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2EX3
 
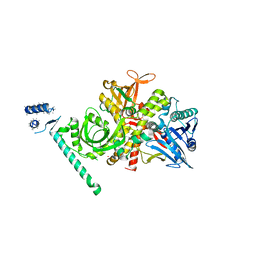 | | Bacteriophage phi29 DNA polymerase bound to terminal protein | | Descriptor: | DNA polymerase, DNA terminal protein, LEAD (II) ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Embo J., 25, 2006
|
|
1XHX
 
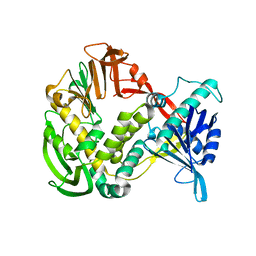 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1XHZ
 
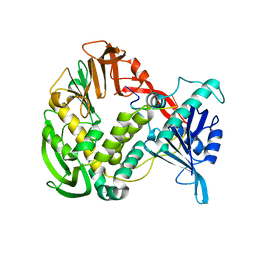 | | Phi29 DNA polymerase, orthorhombic crystal form, ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', DNA polymerase | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1YGM
 
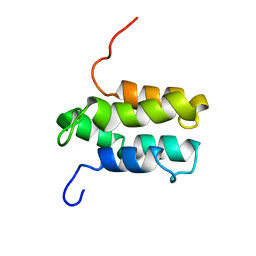 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
4A37
 
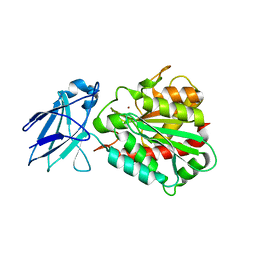 | | Metallo-carboxypeptidase from Pseudomonas Aeruginosa | | Descriptor: | METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4A39
 
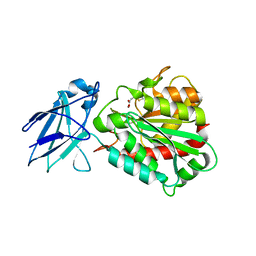 | | Metallo-carboxypeptidase from Pseudomonas Aeruginosa in complex with (2-guanidinoethylmercapto)succinic acid | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
4A38
 
 | | METALLO-CARBOXYPEPTIDASE FROM PSEUDOMONAS AUREGINOSA IN COMPLEX WITH L-BENZYLSUCCINIC ACID | | Descriptor: | L-BENZYLSUCCINIC ACID, METALLO-CARBOXYPEPTIDASE, ZINC ION | | Authors: | Otero, A, Rodriguez de la Vega, M, Tanco, S.M, Lorenzo, J, Aviles, F.X, Reverter, D. | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Structure of a Cytosolic M14 Metallocarboxypeptidase (Ccp) from Pseudomonas Aeruginosa: A Model for Mammalian Ccps.
Faseb J., 26, 2012
|
|
5CGD
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) | | Descriptor: | 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, OLEIC ACID | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
5CGC
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7AZ0
 
 | |
7PQ9
 
 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PA1
 
 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7W
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
