4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U6X
 
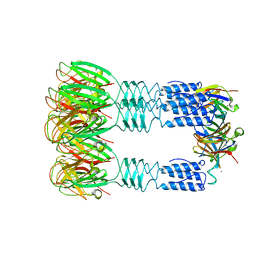 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH8
 
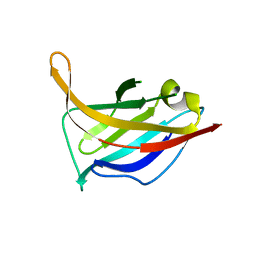 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3D9B
 
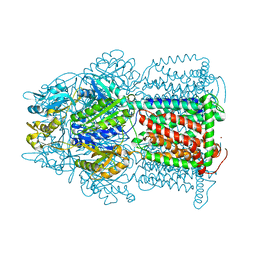 | | Symmetric structure of E. coli AcrB | | Descriptor: | Acriflavine resistance protein B, NICKEL (II) ION | | Authors: | Veesler, D, Blangy, S, Cambillau, C, Sciara, G. | | Deposit date: | 2008-05-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3HG0
 
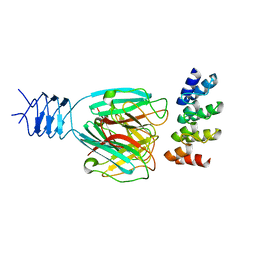 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
3J31
 
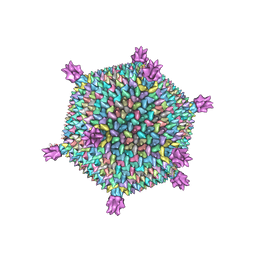 | | Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus | | Descriptor: | A223 penton base, A55 membrane protein, C381 turret protein, ... | | Authors: | Veesler, D, Ng, T.S, Sendamarai, A.K, Eilers, B.J, Lawrence, C.M, Lok, S.M, Young, M.J, Johnson, J.E, Fu, C.-Y. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2X8K
 
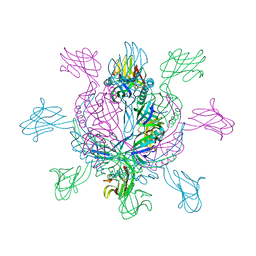 | | Crystal Structure of SPP1 Dit (gp 19.1) Protein, a Paradigm of Hub Adsorption Apparatus in Gram-positive Infecting Phages. | | Descriptor: | HYPOTHETICAL PROTEIN 19.1 | | Authors: | Veesler, D, Robin, G, Lichiere, J, Auzat, I, Tavares, P, Bron, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Bacteriophage Spp1 Distal Tail Protein (Gp 19.1): A Baseplate Hub Paradigm in Gram+ Infecting Phages.
J.Biol.Chem., 285, 2010
|
|
2XF6
 
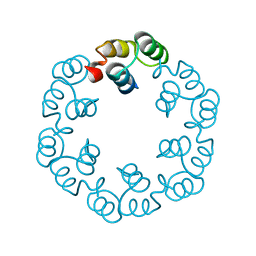 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF5
 
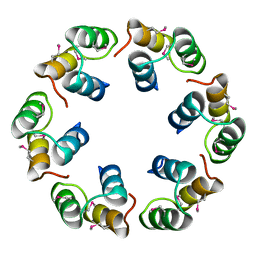 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XC8
 
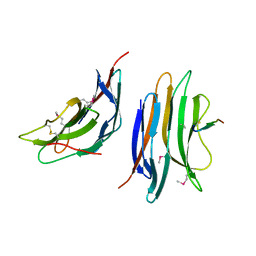 | | Crystal structure of the gene 22 product of the Bacillus subtilis SPP1 phage | | Descriptor: | GENE 22 PRODUCT | | Authors: | Veesler, D, Blangy, S, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp22 Shares Fold Similarity with a Domain of Lactococcal Phage P2 Rbp.
Protein Sci., 19, 2010
|
|
2XF7
 
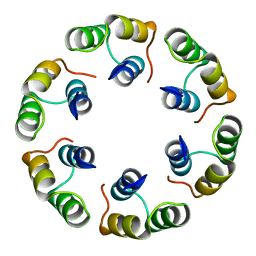 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. High-resolution structure. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
7RA8
 
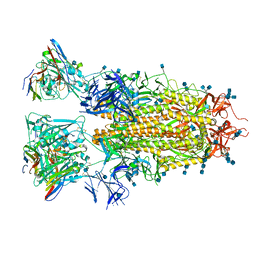 | |
7RAL
 
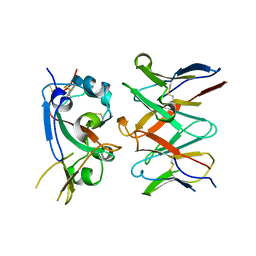 | |
8S9G
 
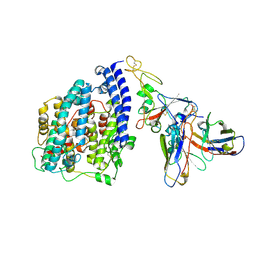 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
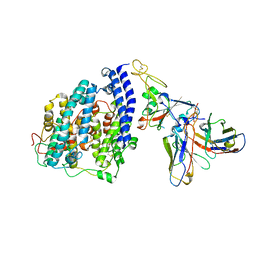 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXB
 
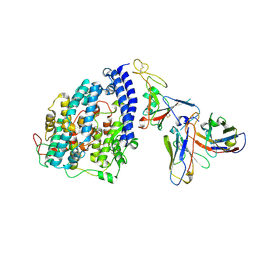 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7N8I
 
 | |
7N8H
 
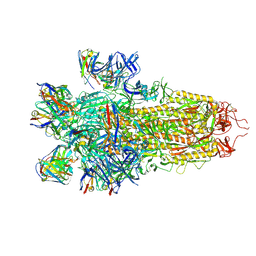 | |
8ERR
 
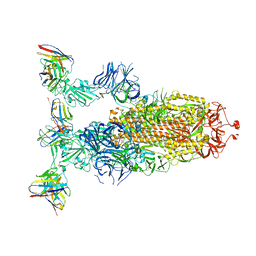 | |
8ERQ
 
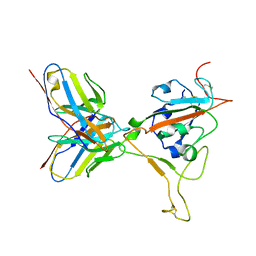 | |
8DYA
 
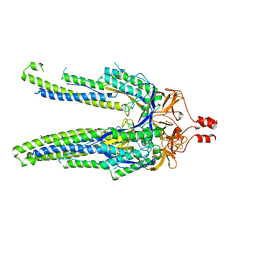 | |
8DF5
 
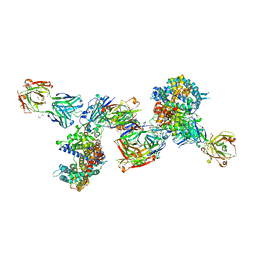 | | SARS-CoV-2 Beta RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McCallum, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.
Science, 377, 2022
|
|
8UR7
 
 | |
8UR5
 
 | |
6X79
 
 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
