2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | Authors: | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
2VDC
 
 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
1LLZ
 
 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: reduced enzyme | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, Ferredoxin-dependent glutamate synthase | | Authors: | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
1LLW
 
 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
1LM1
 
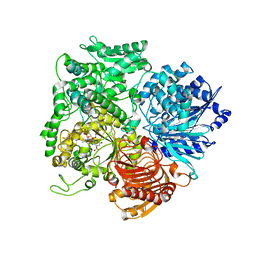 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: native enzyme | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | van Den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
1EA0
 
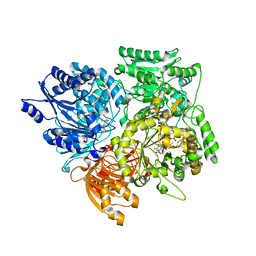 | | Alpha subunit of A. brasilense glutamate synthase | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Binda, C, Bossi, R.T, Vanoni, M.A, Mattevi, A. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-Talk and Ammonia Channeling between Active Centers in the Unexpected Domain Arrangement of Glutamate Synthase
Structure, 8, 2000
|
|
7ZS6
 
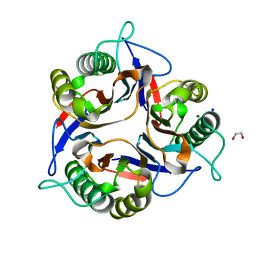 | | Crystal structure of Apis mellifera RidA | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Reactive intermediate deaminase A, ... | | Authors: | Visentin, C, Rizzi, G, Ricagno, S. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
8A8Z
 
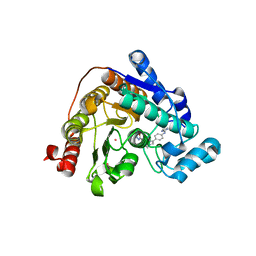 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with in situ enzymatically hydrolyzed DFMO-based ITF5924 | | Descriptor: | 4-[[4-[4-(imidazolidin-2-ylideneamino)phenyl]-1,2,3-triazol-1-yl]methyl]benzohydrazide, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Difluoromethyl-1,3,4-oxadiazoles are slow-binding substrate analog inhibitors of histone deacetylase 6 with unprecedented isotype selectivity.
J.Biol.Chem., 299, 2022
|
|
3JQP
 
 | |
3JQR
 
 | |
3JQQ
 
 | |
6Q9Z
 
 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6S6U
 
 | |
6S6S
 
 | |
6S6X
 
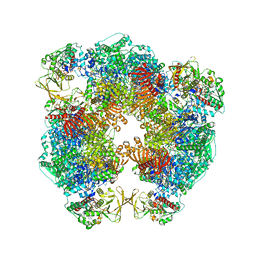 | | Structure of Azospirillum brasilense Glutamate Synthase in a6b6 oligomeric state. | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Camilloni, C, Bolognesi, M. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-11 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures of Azospirillum brasilense Glutamate Synthase in Its Oligomeric Assemblies.
J.Mol.Biol., 431, 2019
|
|
6S6T
 
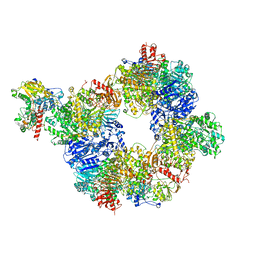 | |
6TCC
 
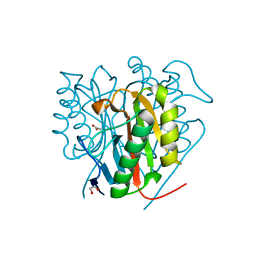 | | Crystal structure of Salmo salar RidA-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
6TCD
 
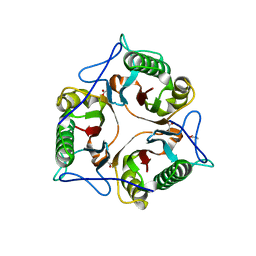 | | Crystal structure of Salmo salar RidA-2 | | Descriptor: | ACETATE ION, Ribonuclease UK114, SULFATE ION | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
1KIF
 
 | | D-AMINO ACID OXIDASE FROM PIG KIDNEY | | Descriptor: | BENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Todone, F, Mattevi, A. | | Deposit date: | 1996-01-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1OFF
 
 | |
1OFD
 
 | |
1OFE
 
 | |
1DDO
 
 | |
