1H12
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1H13
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1H14
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
2R9Z
 
 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
1HCU
 
 | | alpha-1,2-mannosidase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1,2-MANNOSIDASE, CALCIUM ION | | Authors: | Van Petegem, F, Contreras, H, Contreras, R, Van Beeumen, J. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Trichoderma Reesei Alpha-1,2-Mannosidase: Structural Basis for the Cleavage of Four Consecutive Mannose Residues
J.Mol.Biol., 312, 2001
|
|
1H1N
 
 | |
1T0H
 
 | | Crystal structure of the Rattus norvegicus voltage gated calcium channel beta subunit isoform 2a | | Descriptor: | CHLORIDE ION, VOLTAGE-GATED CALCIUM CHANNEL SUBUNIT BETA2A | | Authors: | Van Petegem, F, Clark, K, Chatelain, F, Minor Jr, D. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain.
Nature, 429, 2004
|
|
1T0J
 
 | | Crystal structure of a complex between voltage-gated calcium channel beta2a subunit and a peptide of the alpha1c subunit | | Descriptor: | CHLORIDE ION, Voltage-dependent L-type calcium channel alpha-1C subunit, voltage-gated calcium channel subunit beta2a | | Authors: | Van Petegem, F, Clark, K, Chatelain, F, Minor Jr, D. | | Deposit date: | 2004-04-09 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain.
Nature, 429, 2004
|
|
2BE6
 
 | | 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin 2, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, Chatelain, F.C, Minor Jr, D.L. | | Deposit date: | 2005-10-23 | | Release date: | 2005-11-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
3IM5
 
 | |
3IM6
 
 | |
3IM7
 
 | |
6MM7
 
 | |
6MM5
 
 | |
6MM6
 
 | |
6MM8
 
 | |
4I0Y
 
 | |
7TCI
 
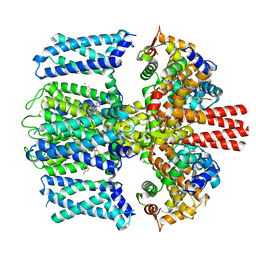 | | Structure of Xenopus KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, CALCIUM ION, Calmodulin-1, ... | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
7TCP
 
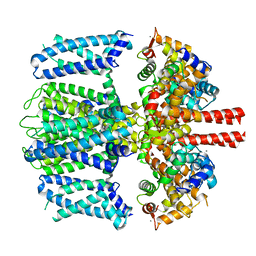 | | Structure of Xenopus KCNQ1-CaM | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
8F5B
 
 | |
5HGC
 
 | | A Serpin structure | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, SULFATE ION, ... | | Authors: | Das, S, Vashchenko, G.V, Van Petegem, F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A Serpin structure
J.Biol.Chem., 2016
|
|
1PG5
 
 | | CRYSTAL STRUCTURE OF THE UNLIGATED (T-STATE) ASPARTATE TRANSCARBAMOYLASE FROM THE EXTREMELY THERMOPHILIC ARCHAEON SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | De Vos, D, Van Petegem, F, Remaut, H, Legrain, C, Glansdorff, N, Van Beeumen, J.J. | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of T State Aspartate Carbamoyltransferase of the Hyperthermophilic Archaeon Sulfolobus acidocaldarius.
J.Mol.Biol., 339, 2004
|
|
6PAL
 
 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
6PYA
 
 | |
