7PIM
 
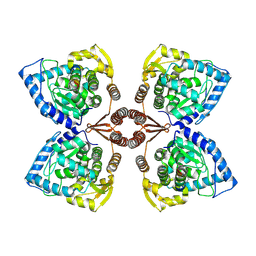 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6QB8
 
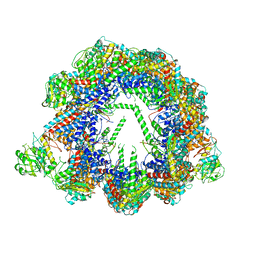 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
1H5W
 
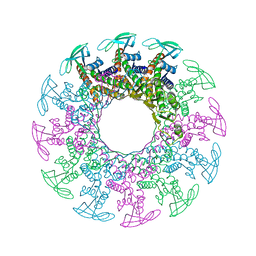 | | 2.1A Bacteriophage Phi-29 Connector | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, UPPER COLLAR PROTEIN | | Authors: | Guasch, A, Pous, J, Ibarra, B, Gomis-Ruth, F.X, Valpuesta, J.M, Sousa, N, Carrascosa, J.L, Coll, M. | | Deposit date: | 2001-05-28 | | Release date: | 2002-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detailed Architecture of a DNA Translocating Machine: The High-Resolution Structure of the Bacteriophage Phi29 Connector Particle.
J.Mol.Biol., 315, 2002
|
|
2XQL
 
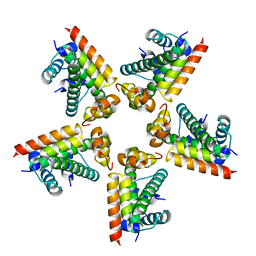 | | Fitting of the H2A-H2B histones in the electron microscopy map of the complex Nucleoplasmin:H2A-H2B histones (1:5). | | Descriptor: | HISTONE H2A-IV, HISTONE H2B 5 | | Authors: | Ramos, I, Martin-Benito, J, Finn, R, Bretana, L, Aloria, K, Arizmendi, J.M, Ausio, J, Muga, A, Valpuesta, J.M, Prado, A. | | Deposit date: | 2010-09-02 | | Release date: | 2010-11-03 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (19.5 Å) | | Cite: | Nucleoplasmin Binds Histone H2A-H2B Dimers Through its Distal Face.
J.Biol.Chem., 285, 2010
|
|
3IZG
 
 | | Bacteriophage T7 prohead shell EM-derived atomic model | | Descriptor: | Major capsid protein 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
2WFS
 
 | | Fitting of influenza virus NP structure into the 9-fold symmetryzed cryoEM reconstruction of an active RNP particle. | | Descriptor: | NUCLEOPROTEIN | | Authors: | Coloma, R, Valpuesta, J.M, Arranz, R, Carrascosa, J.L, Ortin, J, Martin-Benito, J. | | Deposit date: | 2009-04-15 | | Release date: | 2009-07-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The Structure of a Biologically Active Influenza Virus Ribonucleoprotein Complex.
Plos Pathog., 5, 2009
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
2XSM
 
 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3J4A
 
 | | Structure of gp8 connector protein | | Descriptor: | Head-to-tail joining protein | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
3J4B
 
 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | Descriptor: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-27 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZZU
 
 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZN2
 
 | | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | | Descriptor: | FE (III) ION, L-DOPAMINE, SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
4BBL
 
 | | Cryo-electron microscopy reconstruction of the helical part of influenza A virus ribonucleoprotein isolated from virions. | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Arranz, R, Coloma, R, Chichon, F.J, Conesa, J.J, Carrascosa, J.L, Valpuesta, J.M, Ortin, J, Martin-Benito, J. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-05 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of Native Influenza Virion Ribonucleoproteins
Science, 338, 2012
|
|
4BIJ
 
 | | Threading model of T7 large terminase | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
4BIL
 
 | | Threading model of the T7 large terminase within the gp8gp19 complex | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
7ZHS
 
 | | 3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry | | Descriptor: | Ubiquitin-like protein SMT3,DnaJ homolog subfamily A member 2, ZINC ION | | Authors: | Cuellar, J, Velasco-Carneros, L, Santiago, C, Martin-Benito, J, Valpuesta, J, Muga, A. | | Deposit date: | 2022-04-07 | | Release date: | 2023-07-26 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The self-association equilibrium of DNAJA2 regulates its interaction with unfolded substrate proteins and with Hsc70.
Nat Commun, 14, 2023
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
3C7N
 
 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6QIZ
 
 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
4ICV
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal form B | | Descriptor: | PRASEODYMIUM ION, Tubulin-specific chaperone E | | Authors: | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | Deposit date: | 2012-12-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
4ICU
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal from A | | Descriptor: | Tubulin-specific chaperone E | | Authors: | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | Deposit date: | 2012-12-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
6YTU
 
 | | Atomic-resolution structure of the coiled-coil dimerisation domain of human Arc | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Activity-regulated cytoskeleton-associated protein, CHLORIDE ION | | Authors: | Hallin, E.I, Touma, C, Bramham, C.R, Kursula, P. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif.
Febs J., 288, 2021
|
|
