2O5G
 
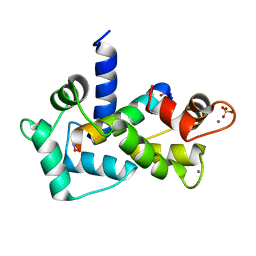 | | Calmodulin-smooth muscle light chain kinase peptide complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Valentine, K.G, Ng, H.L, Schneeweis, J.K, Kranz, J.K, Frederick, K.K, Alber, T, Wand, A.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ultrahigh resolution crystal structure of calmodulin-smooth muscle light kinase peptide complex
To be Published
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2021-10-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
2MI7
 
 | | Solution NMR structure of alpha3Y | | Descriptor: | de novo protein a3Y | | Authors: | Glover, S.D, Jorge, C, Liang, L, Valentine, K.G, Hammarstrom, L, Tommos, C. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Photochemical tyrosine oxidation in the structurally well-defined alpha 3Y protein: proton-coupled electron transfer and a long-lived tyrosine radical.
J.Am.Chem.Soc., 136, 2014
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2LXY
 
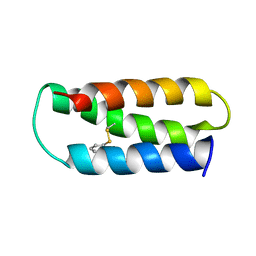 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
2KV9
 
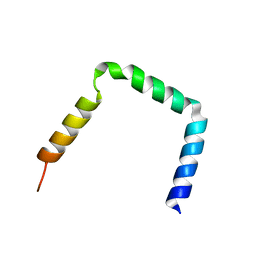 | | Integrin beta3 subunit in a disulfide linked alphaIIb-beta3 cytosolic domain | | Descriptor: | Integrin beta-3 | | Authors: | Metcalf, D.G, Kielec, J.M, Valentine, K.G, Wand, A, Bennett, J.S, William, D.F, Moore, D.T, Molnar, K. | | Deposit date: | 2010-03-10 | | Release date: | 2011-01-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the {alpha}IIb{beta}3 cytoplasmic interaction suggests a mechanism for integrin regulation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2O60
 
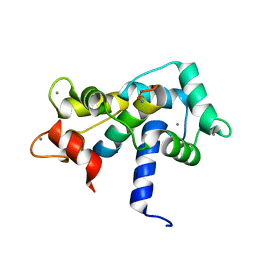 | |
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BZB
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2LEH
 
 | | Solution structure of the core SMN-Gemin2 complex | | Descriptor: | Survival motor neuron protein, Survival of motor neuron protein-interacting protein 1 | | Authors: | Sarachan, K.L, Valentine, K, Gupta, K, Moorman, V, Gledhill, J, Bernens, M, Tommos, C, Wand, A.J, Van Duyne, G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-06-27 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core SMN-Gemin2 complex.
Biochem.J., 445, 2012
|
|
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
