1YRL
 
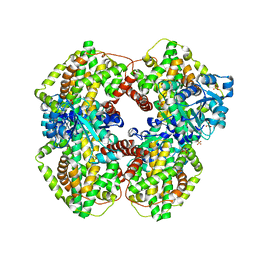 | | Escherichia coli ketol-acid reductoisomerase | | Descriptor: | Ketol-acid reductoisomerase, SULFATE ION | | Authors: | Tyagi, R, Duquerroy, S, Navaza, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-02-04 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a bacterial class II ketol-acid reductoisomerase: domain conservation and evolution
Protein Sci., 14, 2005
|
|
3BE3
 
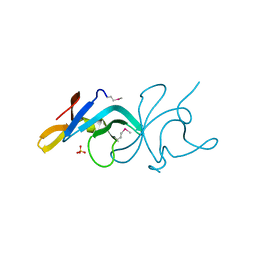 | |
2GOK
 
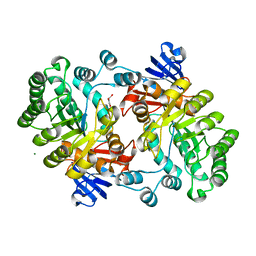 | | Crystal structure of the imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution | | Descriptor: | CHLORIDE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Tyagi, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-13 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
2I6E
 
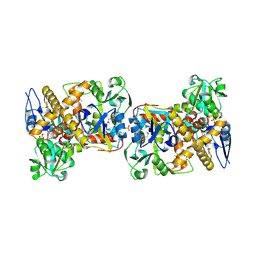 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
2IOJ
 
 | |
2NXO
 
 | |
2OOF
 
 | |
2PBE
 
 | |
2PUZ
 
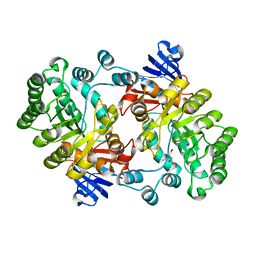 | | Crystal structure of Imidazolonepropionase from Agrobacterium tumefaciens with bound product N-formimino-L-Glutamate | | Descriptor: | CHLORIDE ION, FE (III) ION, Imidazolonepropionase, ... | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
2Q09
 
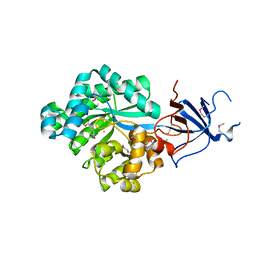 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2QVG
 
 | |
2RK9
 
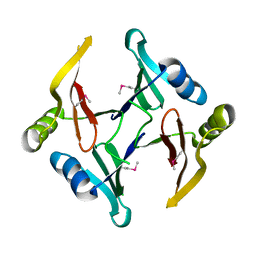 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01 | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Tyagi, R, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01.
To be Published
|
|
1CNM
 
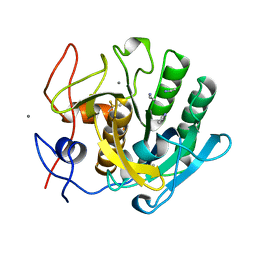 | | ENHANCEMENT OF CATALYTIC EFFICIENCY OF PROTEINASE K THROUGH EXPOSURE TO ANHYDROUS ORGANIC SOLVENT AT 70 DEGREES CELSIUS | | Descriptor: | ACETONITRILE, CALCIUM ION, PROTEIN (PROTEINASE K) | | Authors: | Gupta, M.N, Tyagi, R, Sharma, S, Karthikeyan, S, Singh, T.P. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enhancement of catalytic efficiency of enzymes through exposure to anhydrous organic solvent at 70 degrees C. Three-dimensional structure of a treated serine proteinase at 2.2 A resolution.
Proteins, 39, 2000
|
|
5H2U
 
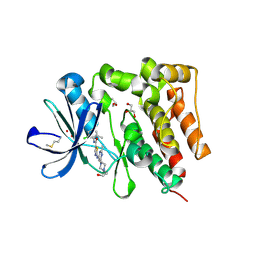 | | Crystal structure of PTK6 Kinase Domain complexed with Dasatinib | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5DA3
 
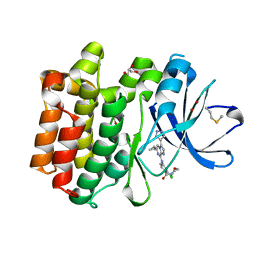 | | Crystal structure of PTK6 Kinase domain with inhibitor | | Descriptor: | (2-chloro-4-{[6-cyclopropyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}phenyl)(morpholin-4-yl)methanone, GLYCEROL, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Battula, S.K, Vadivelu, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5D7V
 
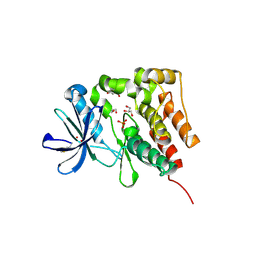 | | Crystal structure of PTK6 kinase domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-14 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the kinase domain of human protein tyrosine kinase 6 (PTK6) at 2.33 angstrom resolution
Biochem.Biophys.Res.Commun., 478, 2016
|
|
3RL3
 
 | | Rat metallophosphodiesterase MPPED2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3RL5
 
 | | Rat metallophosphodiesterase MPPED2 H67R Mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Metallophosphoesterase MPPED2, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3RL4
 
 | | Rat metallophosphodiesterase MPPED2 G252H Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Unique utilization of a phosphoprotein phosphatase fold by a mammalian phosphodiesterase associated with WAGR syndrome.
J.Mol.Biol., 412, 2011
|
|
3IB7
 
 | | Crystal structure of full length Rv0805 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability
J.Biol.Chem., 284, 2009
|
|
3IB8
 
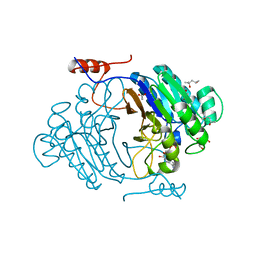 | | Crystal structure of full length Rv0805 in complex with 5'-AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability
J.Biol.Chem., 284, 2009
|
|
4J47
 
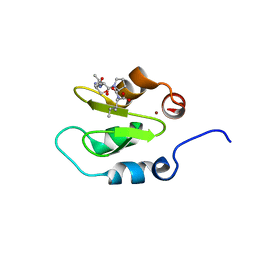 | | Crystal structure of XIAP-BIR2 domain with SVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (SER-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J3Y
 
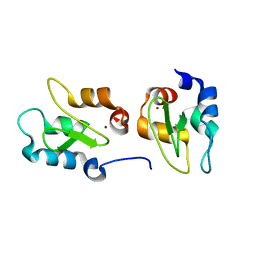 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J48
 
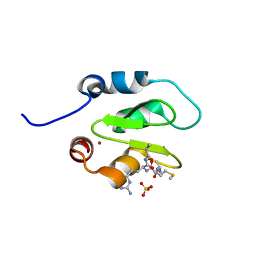 | | Crystal structure of XIAP-BIR2 domain with AMRV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, GLYCEROL, PEPTIDE (ALA-MET-ARG-VAL), ... | | Authors: | Gosu, R. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J46
 
 | | Crystal structure of XIAP-BIR2 domain with AVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
