6I8X
 
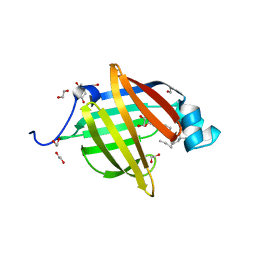 | | As-p18, an extracellular fatty acid binding protein | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein homolog, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Gabrielsen, M, Riboldi-Tunnicliffe, A, Ibanez-Shimabukuro, M, Smith, B.O. | | Deposit date: | 2018-11-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
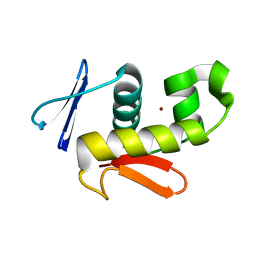
6CQG
 
 | |
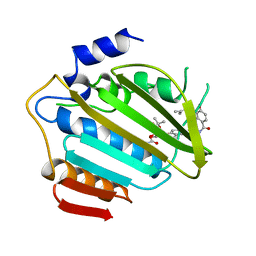
1AJ6
 
 | |
3HRA
 
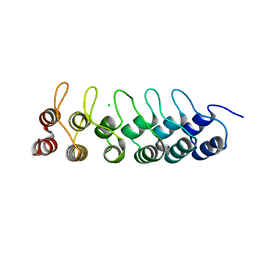 | |
3I75
 
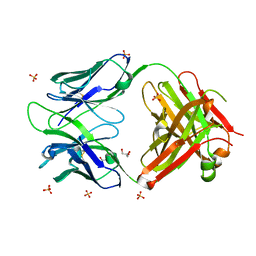 | | Antibody Structure | | Descriptor: | Antibody heavy chain, Antibody light chain, CHLORIDE ION, ... | | Authors: | Isaacs, N.W, Riboldi-Tunnicliffe, A. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody Structure
To be Published
|
|
1FD9
 
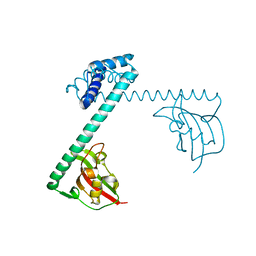 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
2WOZ
 
 | | The novel beta-propeller of the BTB-Kelch protein Krp1 provides the binding site for Lasp-1 that is necessary for pseudopodia extension | | Descriptor: | KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 10 | | Authors: | Gray, C.H, McGarry, L.C, Spence, H.J, Riboldi-Tunnicliffe, A, Ozanne, B.W. | | Deposit date: | 2009-07-31 | | Release date: | 2009-09-01 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel beta-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension.
J. Biol. Chem., 284, 2009
|
|
1NXP
 
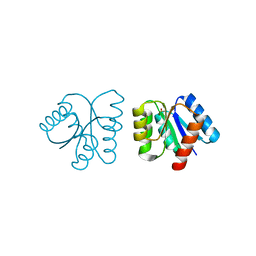 | | MicArec pH4.5 | | Descriptor: | DNA-binding response regulator, PHOSPHONIC ACID | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXO
 
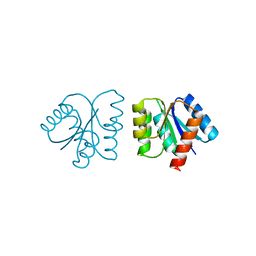 | | MicArec pH7.0 | | Descriptor: | DNA-binding response regulator | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXT
 
 | | MicArec pH 4.0 | | Descriptor: | DNA-binding response regulator, XENON | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXW
 
 | | MicArec pH 5.1 | | Descriptor: | ACETIC ACID, DNA-binding response regulator | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
6EB7
 
 | |
6EBR
 
 | |
6EBB
 
 | |
1YIV
 
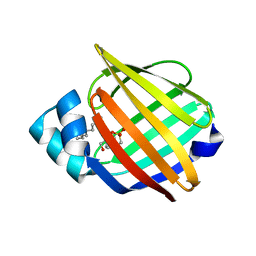 | | Structure of myelin P2 protein from Equine spinal cord | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Myelin P2 protein | | Authors: | Hunter, D.J.B, MacMaster, R, Rozak, A.W, Riboldi-Tunnicliffe, A, Grifiths, I.R, Freer, A.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of myelin P2 protein from equine spinal cord.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NXV
 
 | |
1NXS
 
 | |
1NXX
 
 | |
7KGS
 
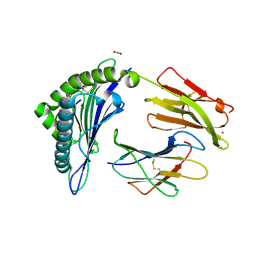 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N138-146 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGT
 
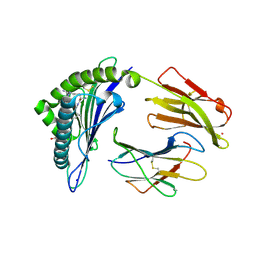 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGR
 
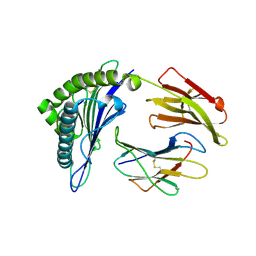 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N159-167 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nucleoprotein | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGQ
 
 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N222-230 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGP
 
 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGO
 
 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
2A9P
 
 | | Medium Resolution BeF3 bound RR02-rec | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MANGANESE (II) ION, Response regulator | | Authors: | Riboldi-Tunnicliffe, A, Isaacs, N.W, Mitchell, T.J. | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of an activated YycF homologue, the essential response regulator from S.pneumoniae in complex with BeF3 and the effect of pH on BeF3 binding, possible phosphate in the active site.
TO BE PUBLISHED
|
|
