3R9X
 
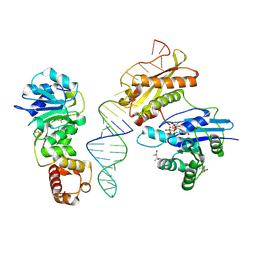 | | Crystal structure of Era in complex with MgGDPNP, nucleotides 1506-1542 of 16S ribosomal RNA, and KsgA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, GTPase Era, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2011-03-26 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R9W
 
 | |
3FTD
 
 | |
3FTF
 
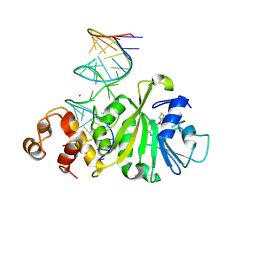 | | Crystal structure of A. aeolicus KsgA in complex with RNA and SAH | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase, POTASSIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
3FTC
 
 | |
3FTE
 
 | | Crystal structure of A. aeolicus KsgA in complex with RNA | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
3IEU
 
 | | Crystal Structure of ERA in Complex with GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GTP-binding protein era, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IEV
 
 | | Crystal Structure of ERA in Complex with MgGNP and the 3' End of 16S rRNA | | Descriptor: | 5'-R(P*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3', GTP-binding protein era, MAGNESIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
5C6W
 
 | | anti-CXCL13 scFv - E10 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
5C2B
 
 | | anti-CXCL13 parental scFv - 3B4 | | Descriptor: | CHLORIDE ION, scFv 3B4 | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-15 | | Release date: | 2015-11-04 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.4049 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
5CBA
 
 | | 3B4 in complex with CXCL13 - 3B4-CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, 3b4 heavy chain, 3b4 light chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
2PCX
 
 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4GLR
 
 | | Structure of the anti-ptau Fab (pT231/pS235_1) in complex with phosphoepitope pT231/pS235 | | Descriptor: | PHOSPHATE ION, anti-ptau heavy chain, anti-ptau light chain, ... | | Authors: | Tu, C, Mosyak, L, Bard, J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Ultra-specific Avian Antibody to Phosphorylated Tau Protein Reveals a Unique Mechanism for Phosphoepitope Recognition.
J.Biol.Chem., 287, 2012
|
|
1KEQ
 
 | | Crystal Structure of F65A/Y131C Carbonic Anhydrase V, covalently modified with 4-chloromethylimidazole | | Descriptor: | 4-METHYLIMIDAZOLE, ACETIC ACID, F65A/Y131C-MI Carbonic Anhydrase V, ... | | Authors: | Jude, K.M, Wright, S.K, Tu, C, Silverman, D.N, Viola, R.E, Christianson, D.W. | | Deposit date: | 2001-11-16 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle.
Biochemistry, 41, 2002
|
|
1Z97
 
 | | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1Z93
 
 | | Human Carbonic Anhydrase III:Structural and Kinetic study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1TB0
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II.
Biochemistry, 44, 2005
|
|
3L14
 
 | | Human Carbonic Anhydrase II complexed with Althiazide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Quirit, J.G, Robbins, A, Genis, C, Tu, C, Silverman, D.N, McKenna, R. | | Deposit date: | 2009-12-10 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The Promiscuous Nature of Althiazide in Adducts with CA II and CA IX Mimic
To be Published
|
|
2ESF
 
 | | Identification of a Novel Non-Catalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Gareiss, P.C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase.
Biochemistry, 45, 2006
|
|
2FNM
 
 | | Activation of human carbonic anhdyrase II by exogenous proton donors | | Descriptor: | ZINC ION, carbonic anhydrase 2 | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
2FNK
 
 | | Activation of Human Carbonic Anhydrase II by exogenous proton donors | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
2FNN
 
 | | Activation of human carbonic anhydrase II by exogenous proton donors | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic Anhydrase 2, ZINC ION | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
1YO1
 
 | | Proton Transfer from His200 in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Bhatt, D, Tu, C, Fisher, S.Z, Hernandez Prada, J.A, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-01-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proton transfer in a Thr200His mutant of human carbonic anhydrase II
Proteins, 61, 2005
|
|
1YO0
 
 | | Proton Transfer from His200 in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Bhatt, D, Tu, C, Fisher, S.Z, Hernandez Prada, J.A, McKenna, R, Silverman, D.N. | | Deposit date: | 2005-01-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton transfer in a Thr200His mutant of human carbonic anhydrase II
Proteins, 61, 2005
|
|
