1A1S
 
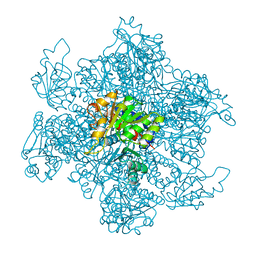 | | ORNITHINE CARBAMOYLTRANSFERASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE | | Authors: | Villeret, V, Clantin, B, Tricot, C, Legrain, C, Roovers, M, Stalon, V, Glansdorff, N, Van Beeumen, J. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DXH
 
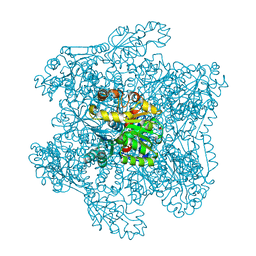 | | Catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, SULFATE ION | | Authors: | Sainz, G, Vicat, J, Kahn, R, Duee, E, Tricot, C, Stalon, V, Dideberg, O. | | Deposit date: | 2000-01-05 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Allosteric Active Form of Catabolic Ornithine Carbamoyltransferase from Pseudomonas Aeruginosa
To be Published
|
|
1HX3
 
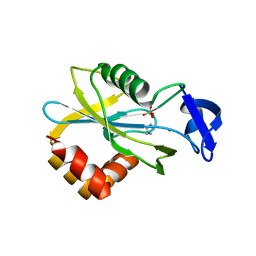 | | CRYSTAL STRUCTURE OF E.COLI ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | IMIDAZOLE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-11 | | Release date: | 2001-07-11 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
1HZT
 
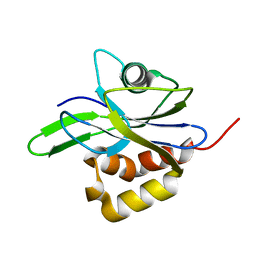 | | CRYSTAL STRUCTURE OF METAL-FREE ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
2FCA
 
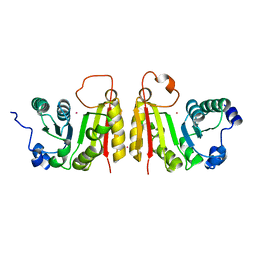 | | The structure of BsTrmB | | Descriptor: | POTASSIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zegers, I, Van Vliet, F, Bujnicki, J, Kosinski, J, Gigot, D, Droogmans, L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase.
Nucleic Acids Res., 34, 2006
|
|
3LGA
 
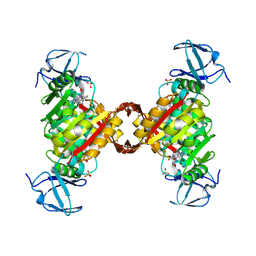 | |
1NFS
 
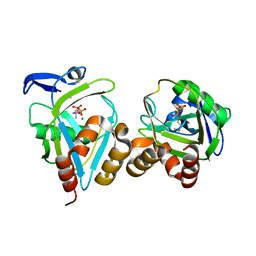 | |
1NFZ
 
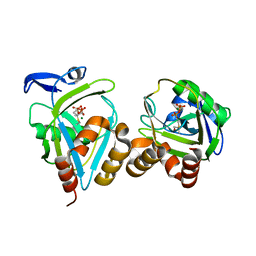 | |
1ORT
 
 | | ORNITHINE TRANSCARBAMOYLASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ORNITHINE TRANSCARBAMOYLASE | | Authors: | Villeret, V, Dideberg, O. | | Deposit date: | 1995-08-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3LHD
 
 | | Crystal structure of P. abyssi tRNA m1A58 methyltransferase in complex with S-adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, putative | | Authors: | Guelorget, A, Golinelli-Pimpaneau, B, Wouters, J, Barbey, C. | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase.
Nucleic Acids Res., 38, 2010
|
|
3MB5
 
 | |
