6Z9M
 
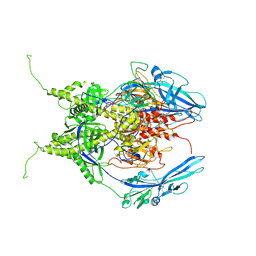 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
3J2T
 
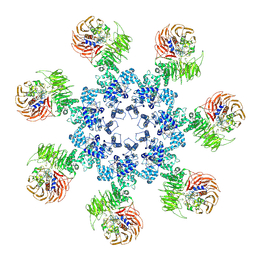 | | An improved model of the human apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Yuan, S, Topf, M, Akey, C.W. | | Deposit date: | 2012-12-23 | | Release date: | 2013-04-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Changes in apaf-1 conformation that drive apoptosome assembly.
Biochemistry, 52, 2013
|
|
2X7N
 
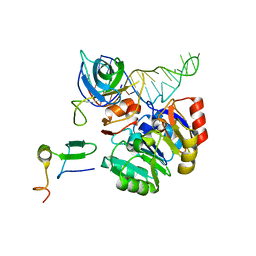 | | Mechanism of eIF6s anti-association activity | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, 60S RIBOSOMAL PROTEIN L24-A, EUKARYOTIC TRANSLATION INITIATION FACTOR 6, ... | | Authors: | Gartmann, M, Blau, M, Armache, J.-P, Mielke, T, Topf, M, Beckmann, R. | | Deposit date: | 2010-03-02 | | Release date: | 2010-03-31 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | Mechanism of Eif6-Mediated Inhibition of Ribosomal Subunit Joining.
J.Biol.Chem., 285, 2010
|
|
7PAG
 
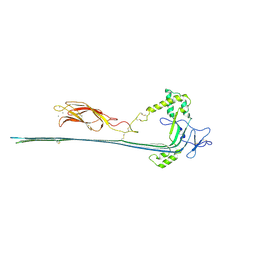 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
1NX4
 
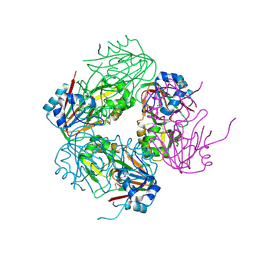 | | The crystal structure of carbapenem synthase (CarC) | | Descriptor: | 2-OXOGLUTARIC ACID, Carbapenem synthase, FE (III) ION | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-08 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
1NX8
 
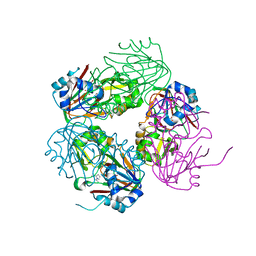 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
4V3N
 
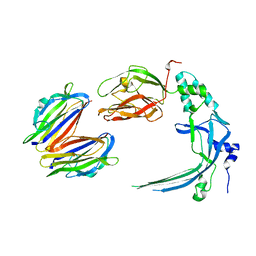 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
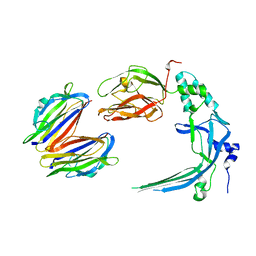 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
3B63
 
 | | Actin filament model in the extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments.
J.Mol.Biol., 375, 2008
|
|
3B5U
 
 | | Actin filament model from extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments
J.Mol.Biol., 375, 2008
|
|
7NB8
 
 | | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-5, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
7NBA
 
 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
4UXT
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN HEAVY CHAIN ISOFORM 5A, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4V7H
 
 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
8B6U
 
 | | Mpf2Ba1 monomer | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez-Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6W
 
 | | Mpf2Ba1 pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6V
 
 | | Mp2Ba1 pre-pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
4V4L
 
 | | Structure of the Drosophila apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK, MAGNESIUM ION | | Authors: | Yuan, S, Topf, M, Akey, C.W, Ludtke, S.J. | | Deposit date: | 2010-10-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the Drosophila apoptosome at 6.9 angstrom resolution
Structure, 19, 2011
|
|
4UXS
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXR
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UY0
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXO
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2018-04-04 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UXP
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
