7W8F
 
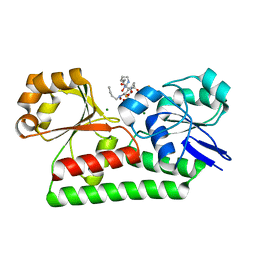 | |
2ZEQ
 
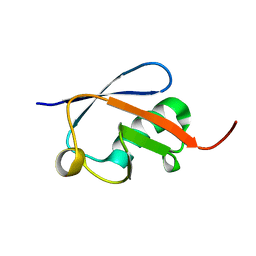 | | Crystal structure of ubiquitin-like domain of murine Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin | | Authors: | Tomoo, K. | | Deposit date: | 2007-12-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin
Biochim.Biophys.Acta, 1784, 2008
|
|
1WKW
 
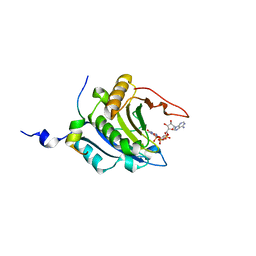 | | Crystal structure of the ternary complex of eIF4E-m7GpppA-4EBP1 peptide | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Matsushita, Y, Fujisaki, H, Shen, X, Miyagawa, H, Kitamura, K, Miura, K, Ishida, T. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods
Biochim.Biophys.Acta, 1753, 2005
|
|
1IPB
 
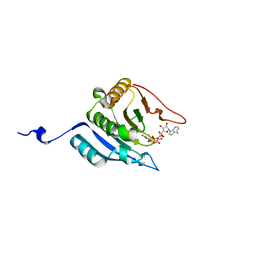 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1IPC
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
3VXC
 
 | |
3VXB
 
 | |
3B1L
 
 | |
3AM7
 
 | | Crystal structure of the ternary complex of eIF4E-M7GTP-4EBP2 peptide | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 2 | | Authors: | Tomoo, K, Fukuyo, A, In, Y, Ishida, T. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural scaffold for eIF4E binding selectivity of 4E-BP isoforms: crystal structure of eIF4E binding region of 4E-BP2 and its comparison with that of 4E-BP1.
J.Pept.Sci., 17, 2011
|
|
1BTP
 
 | | UNIQUE BINDING OF A NOVEL SYNTHETIC INHIBITOR, N-[3-[4-[4-(AMIDINOPHENOXY)-CARBONYL]PHENYL]-2-METHYL-2-PROPENOYL]-N-ALLYLGLYCINE METHANESULFONATE TO BOVINE TRYPSIN, REVEALED BY THE CRYSTAL STRUCTURE OF THE COMPLEX | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Odagaki, Y, Nakai, H, Senokuchi, K, Kawamura, M, Hamanaka, N, Nakamura, M, Tomoo, K, Ishida, T. | | Deposit date: | 1995-08-11 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique binding of a novel synthetic inhibitor, N-[3-[4-[4-(amidinophenoxy)carbonyl]phenyl]-2-methyl-2-propenoyl]- N-allylglycine methanesulfonate, to bovine trypsin, revealed by the crystal structure of the complex.
Biochemistry, 34, 1995
|
|
7VTJ
 
 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
1CWY
 
 | | CRYSTAL STRUCTURE OF AMYLOMALTASE FROM THERMUS AQUATICUS, A GLYCOSYLTRANSFERASE CATALYSING THE PRODUCTION OF LARGE CYCLIC GLUCANS | | Descriptor: | AMYLOMALTASE | | Authors: | Przylas, I, Tomoo, K, Terada, Y, Takaha, T, Fuji, K, Saenger, W, Straeter, N. | | Deposit date: | 1999-08-27 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amylomaltase from thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans.
J.Mol.Biol., 296, 2000
|
|
7FGR
 
 | | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain. | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fab Heavy Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain.
To Be Published
|
|
7FGK
 
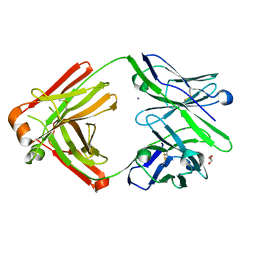 | | The Fab antibody single structure against tau protein. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
7FGJ
 
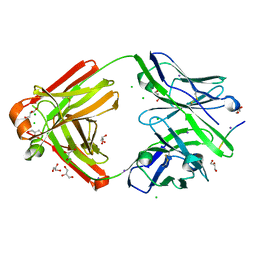 | | The cross-reaction complex structure with VQILNK peptide and the tau antibody's Fab domain. | | Descriptor: | CHLORIDE ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The cross-reaction complex structure with VQILNK peptide and the antibody's Fab domain.
To Be Published
|
|
1VTU
 
 | | Structural characteristics of enantiomorphic DNA: Crystal analysis of racemates of the D(CGCGCG) duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Doi, M, Inoue, M, Tomoo, K, Ishida, T, Ueda, Y, Akagi, M, Urata, H. | | Deposit date: | 1994-08-29 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characteristics of Enantiomorphic DNA: Crystal Analysis of Racemates of the d(CGCGCG) Duplex
J.Am.Chem.Soc., 115, 1993
|
|
1PIP
 
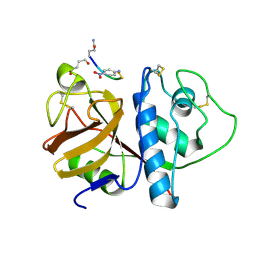 | | CRYSTAL STRUCTURE OF PAPAIN-SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE COMPLEX AT 1.7 ANGSTROMS RESOLUTION: NONCOVALENT BINDING MODE OF A COMMON SEQUENCE OF ENDOGENOUS THIOL PROTEASE INHIBITORS | | Descriptor: | Papain, SUCCINYL-GLN-VAL-VAL-ALA-ALA-P-NITROANILIDE | | Authors: | Yamamoto, A, Tomoo, K, Doi, M, Ohishi, H, Inoue, M, Ishida, T, Yamamoto, D, Tsuboi, S, Okamoto, H, Okada, Y. | | Deposit date: | 1992-10-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papain-succinyl-Gln-Val-Val-Ala-Ala-p-nitroanilide complex at 1.7-A resolution: noncovalent binding mode of a common sequence of endogenous thiol protease inhibitors.
Biochemistry, 31, 1992
|
|
6LRA
 
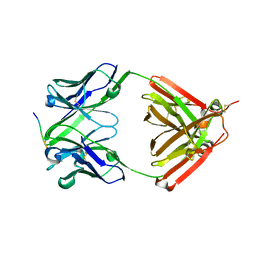 | | The complex structure of PHF core domain peptide of tau and antibody's Fab domain. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, VQIINK | | Authors: | Tomohiro, T, Kouki, S, Tomohiro, S, Takahiro, T, Katsushiro, M, Yasuko, I, Katsuhiko, M, Taizo, T, Toshimitsu, I, Koji, T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the human tau PHF core domain VQIINK complexed with the Fab domain of monoclonal antibody Tau2r3.
Febs Lett., 2020
|
|
1QDQ
 
 | | X-RAY CRYSTAL STRUCTURE OF BOVINE CATHEPSIN B-CA074 COMPLEX | | Descriptor: | CATHEPSIN B, [PROPYLAMINO-3-HYDROXY-BUTAN-1,4-DIONYL]-ISOLEUCYL-PROLINE | | Authors: | Yamamoto, A. | | Deposit date: | 1999-07-10 | | Release date: | 2000-07-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex.
J.Biochem.(Tokyo), 127, 2000
|
|
5JZQ
 
 | |
1PE6
 
 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
1PPP
 
 | |
2DC6
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA073 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-28 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC7
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA042 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-THREONYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCC
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA077 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, CATHEPSIN B, GLYCEROL, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
