7N0X
 
 | | Rhesusized RV144 DH827 Fab bound to HIV-1 Env V2 peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycoprotein 120, Rhesusized RV144 DH827 heavy chain Fab fragment, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-05-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Fc-Effector Function of Rhesusized Variants of Human Anti-HIV-1 IgG1s.
Front Immunol, 12, 2021
|
|
7N8Q
 
 | |
1MSV
 
 | | The S68A S-adenosylmethionine decarboxylase proenzyme processing mutant. | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Tolbert, W.D, Zhang, Y, Bennett, E.M, Cottet, S.E, Ekstrom, J.L, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of Human S-Adenosylmethionine
Decarboxylase Proenzyme Processing as Revealed by the
Structure of the S68A Mutant.
Biochemistry, 42, 2003
|
|
7KCZ
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) IGG1 FC FRAGMENT- FC-GAMMA RECEPTOR III COMPLEX V158 MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
7KJN
 
 | |
7KJM
 
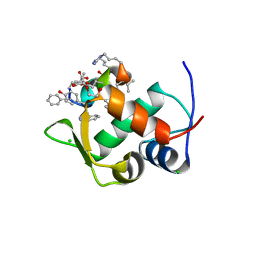 | |
3HMS
 
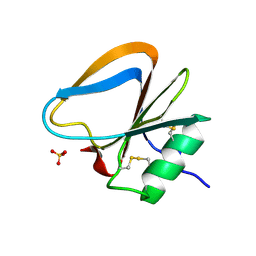 | |
3HMT
 
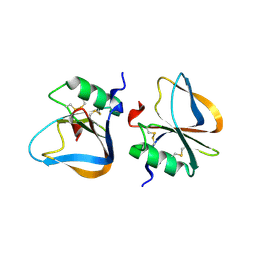 | |
3HMR
 
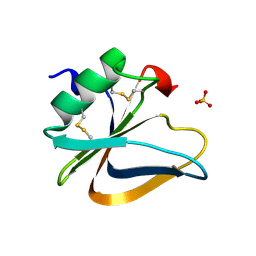 | |
3HN4
 
 | |
2QJ2
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QJ4
 
 | | A Mechanistic Basis for Converting a Receptor Tyrosine Kinase Agonist to an Antagonist | | Descriptor: | Hepatocyte growth factor, SULFATE ION | | Authors: | Tolbert, W.D, Daugherty, J, Gao, C.-F, Xe, Q, Miranti, C, Gherardi, E, Vande Woude, G, Xu, H.E. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6OZ4
 
 | |
6OZ2
 
 | |
4YC2
 
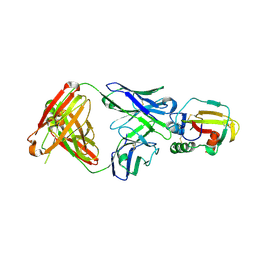 | |
4YBL
 
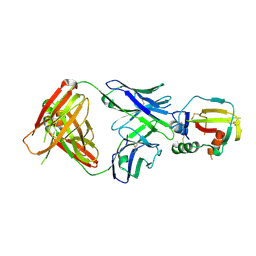 | |
1I7B
 
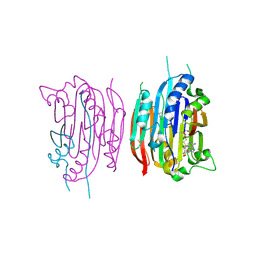 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND S-ADENOSYLMETHIONINE METHYL ESTER | | Descriptor: | 1,4-DIAMINOBUTANE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, S-ADENOSYLMETHIONINE DECARBOXYLASE BETA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I72
 
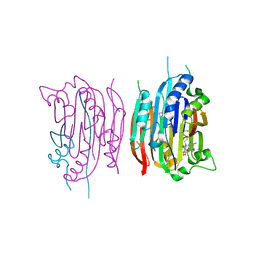 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-07 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7C
 
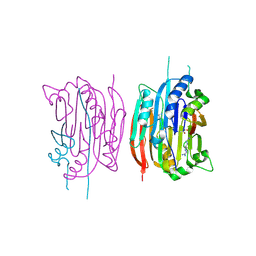 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH METHYLGLYOXAL BIS-(GUANYLHYDRAZONE) | | Descriptor: | 1,4-DIAMINOBUTANE, METHYLGLYOXAL BIS-(GUANYLHYDRAZONE), S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1I7M
 
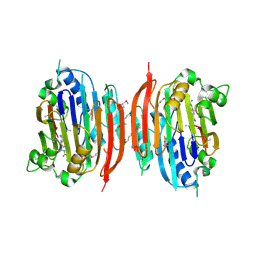 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE | | Descriptor: | 1,4-DIAMINOBUTANE, 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
4FZE
 
 | |
6OFI
 
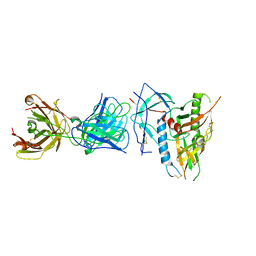 | | CRYSTAL STRUCTURE OF the RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH55 Fab heavy chain, CH55 Fab light chain, ... | | Authors: | Tolbert, W.D, Yan, F, Van, V, Pazgier, M. | | Deposit date: | 2019-03-29 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6OEJ
 
 | |
6OZ3
 
 | |
6W4M
 
 | | CRYSTAL STRUCTURE OF THE ADCC-POTENT, WEAKLY NEUTRALIZING HIV ENV CO-RECEPTOR BINDING SITE ANTIBODY N12-I2 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 AND M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HIV ANTIBODY N12-I2 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY N12-I2 FAB LIGHT CHAIN, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Defining rules governing recognition and Fc-mediated effector functions to the HIV-1 co-receptor binding site.
Bmc Biol., 18, 2020
|
|
