1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | Deposit date: | 1996-12-12 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
1QC1
 
 | |
1QP5
 
 | | BASE-PAIRING SHIFT IN A DODECAMER CONTAINING A (CA)N TRACT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Timsit, Y, Vilbois, E, Moras, D. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Base-pairing shift in the major groove of (CA)n tracts by B-DNA crystal structures.
Nature, 354, 1991
|
|
2GHJ
 
 | | Crystal structure of folded and partially unfolded forms of Aquifex aeolicus ribosomal protein L20 | | Descriptor: | 50S ribosomal protein L20, SULFATE ION | | Authors: | Timsit, Y, Allemand, F, Chiaruttini, C, Springer, M. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-18 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Coexistence of two protein folding states in the crystal structure of ribosomal protein L20
Embo Rep., 7, 2006
|
|
2R22
 
 | |
2R20
 
 | |
2R21
 
 | |
2R1S
 
 | |
330D
 
 | |
329D
 
 | |
384D
 
 | |
382D
 
 | |
383D
 
 | |
8D73
 
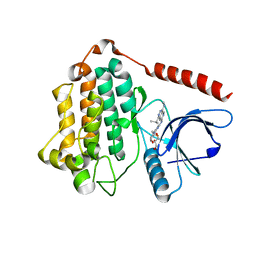 | | Crystal Structure of EGFR LRTM with compound 7 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[4-(methylamino)-1-(propan-2-yl)pyrido[3,4-d]pyridazin-7-yl]amino}pyrimidin-2-yl)piperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
8D76
 
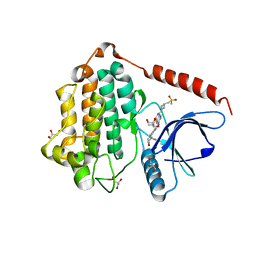 | | Crystal Structure of EGFR LRTM with compound 24 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[8-{3-[(methanesulfonyl)methyl]azetidin-1-yl}-5-(propan-2-yl)-2,7-naphthyridin-3-yl]amino}pyrimidin-2-yl)-3-methylpiperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
