1AO5
 
 | | MOUSE GLANDULAR KALLIKREIN-13 (PRORENIN CONVERTING ENZYME) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLANDULAR KALLIKREIN-13 | | Authors: | Timm, D.E. | | Deposit date: | 1997-07-16 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the mouse glandular kallikrein-13 (prorenin converting enzyme)
Protein Sci., 6, 1997
|
|
3RN8
 
 | |
3RNN
 
 | |
1QCN
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1QQJ
 
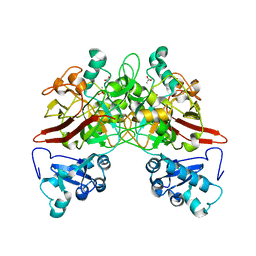 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1QCO
 
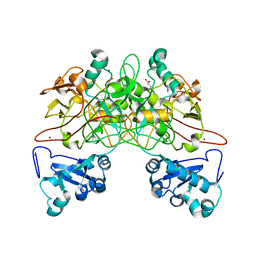 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1I04
 
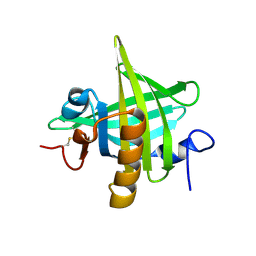 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN-I FROM MOUSE LIVER | | Descriptor: | MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of pheromone binding to mouse major urinary protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I06
 
 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH SEC-BUTYL-THIAZOLINE | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1I05
 
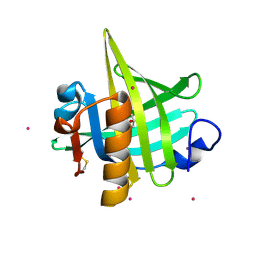 | | CRYSTAL STRUCTURE OF MOUSE MAJOR URINARY PROTEIN (MUP-I) COMPLEXED WITH HYDROXY-METHYL-HEPTANONE | | Descriptor: | 6-HYDROXY-6-METHYL-HEPTAN-3-ONE, CADMIUM ION, MAJOR URINARY PROTEIN I | | Authors: | Timm, D.E, Baker, L.J, Mueller, H, Zidek, L, Novotny, M.V. | | Deposit date: | 2001-01-28 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Pheromone Binding to Mouse Major Urinary Protein (MUP-I)
Protein Sci., 10, 2001
|
|
1IG3
 
 | | Mouse Thiamin Pyrophosphokinase Complexed with Thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, SULFATE ION, thiamin pyrophosphokinase | | Authors: | Timm, D.E, Liu, J, Baker, L.-J, Harris, R.A. | | Deposit date: | 2001-04-16 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamin pyrophosphokinase.
J.Mol.Biol., 310, 2001
|
|
4X7I
 
 | | Crystal Structure of BACE with amino thiazine inhibitor LY2886721 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(4aS,7aS)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Timm, D.E. | | Deposit date: | 2014-12-09 | | Release date: | 2014-12-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Potent BACE1 Inhibitor LY2886721 Elicits Robust Central A beta Pharmacodynamic Responses in Mice, Dogs, and Humans.
J.Neurosci., 35, 2015
|
|
2OGZ
 
 | | Crystal structure of DPP-IV complexed with Lilly aryl ketone inhibitor | | Descriptor: | 4-[(3R)-3-{[2-(4-FLUOROPHENYL)-2-OXOETHYL]AMINO}BUTYL]BENZAMIDE, Dipeptidyl peptidase | | Authors: | Timm, D.E. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of non-covalent dipeptidyl peptidase IV inhibitors which induce a conformational change in the active site.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4YBI
 
 | | Crystal structure of BACE with amino thiazine inhibitor LY2811376 | | Descriptor: | (4S)-4-[2,4-difluoro-5-(pyrimidin-5-yl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor.
J.Neurosci., 31, 2011
|
|
4ZSM
 
 | | BACE crystal structure with bicyclic aminothiazine fragment | | Descriptor: | (4aS,8aR)-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSQ
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4S,4aS,6S,8aR)-10-aminohexahydro-3H-4,8a-(epithiomethenoazeno)isochromen-6(1H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSR
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, N-[(4aS,6S,8aR)-2-amino-5,6,7,8-tetrahydro-4a,8a-(methanooxymethano)-3,1-benzothiazin-6(4H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSP
 
 | | BACE crystal structure with bicyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4aS,6S,8aR)-2-amino-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-6-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
1EY2
 
 | | HUMAN HOMOGENTISATE DIOXYGENASE WITH FE(II) | | Descriptor: | FE (II) ION, HOMOGENTISATE 1,2-DIOXYGENASE | | Authors: | Timm, D.E, Titus, G.P, Penalva, M.A, Mueller, H.A, de Cordoba, S.M. | | Deposit date: | 2000-05-05 | | Release date: | 2000-11-05 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human homogentisate dioxygenase.
Nat.Struct.Biol., 7, 2000
|
|
1EYB
 
 | | CRYSTAL STRUCTURE OF APO HUMAN HOMOGENTISATE DIOXYGENASE | | Descriptor: | HOMOGENTISATE 1,2-DIOXYGENASE | | Authors: | Timm, D.E, Titus, G.P, Penalva, M.A, Mueller, H.A, de Cordoba, S.M. | | Deposit date: | 2000-05-05 | | Release date: | 2000-11-05 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human homogentisate dioxygenase.
Nat.Struct.Biol., 7, 2000
|
|
2HZY
 
 | |
2F17
 
 | | Mouse Thiamin Pyrophosphokinase in a Ternary Complex with Pyrithiamin Pyrophosphate and AMP at 2.5 angstrom | | Descriptor: | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-METHYLPYRIDINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Liu, J.Y, Timm, D.E, Hurley, T.D. | | Deposit date: | 2005-11-14 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrithiamine as a substrate for thiamine pyrophosphokinase
J.Biol.Chem., 281, 2006
|
|
7M05
 
 | |
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVP
 
 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
