1ZXF
 
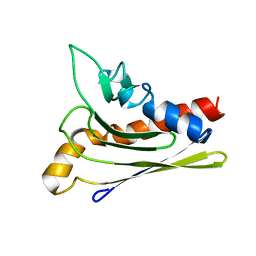 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
7MSY
 
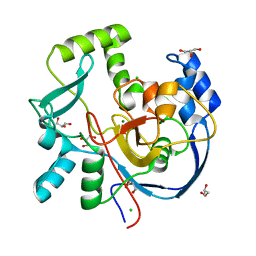 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
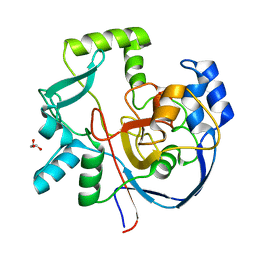 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6N04
 
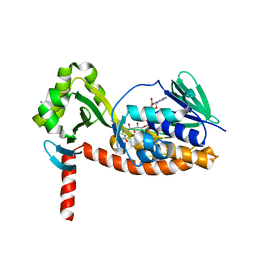 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
5K9M
 
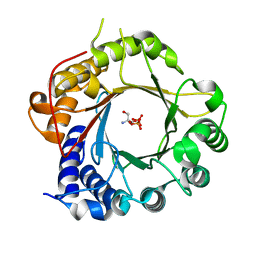 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JXM
 
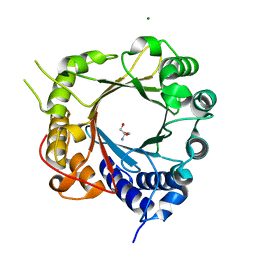 | | Crystal Structure of Prenyltransferase PriB Apo Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5HOQ
 
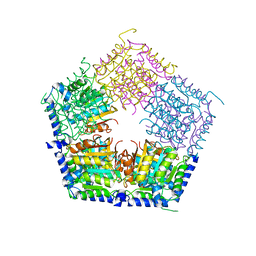 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5INJ
 
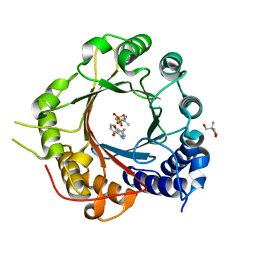 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JR3
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
6D2V
 
 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6D34
 
 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6P9V
 
 | | Crystal Structure of hMAT Mutant K289L | | Descriptor: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Miller, M.D, Xu, W, Huber, T.D, Clinger, J.A, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.
Acs Chem.Biol., 15, 2020
|
|
2GKD
 
 | |
2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
5DU2
 
 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
5EEH
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | Descriptor: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
1IIN
 
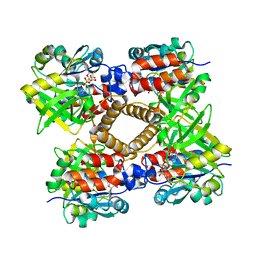 | | thymidylyltransferase complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
1IIM
 
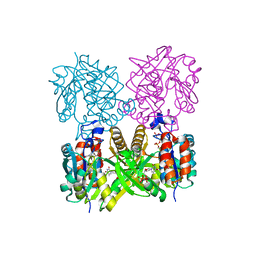 | | thymidylyltransferase complexed with TTP | | Descriptor: | THYMIDINE-5'-TRIPHOSPHATE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
1MP5
 
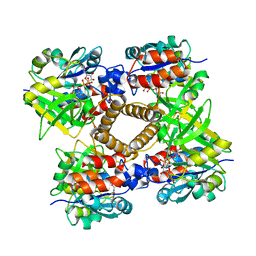 | | Y177F VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, Y177F VARIANT OF S. ENTERICA RmlA BOUND TO UDP-GLUCOSE | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MP4
 
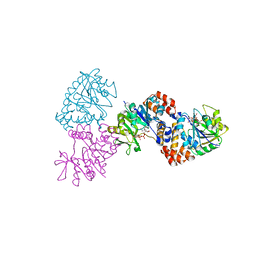 | | W224H VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, W224H Variant of S. Enterica RmlA Bound to UDP-Glucose | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
