2ZNM
 
 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
3DKS
 
 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
6VUB
 
 | |
6VUF
 
 | |
6VUJ
 
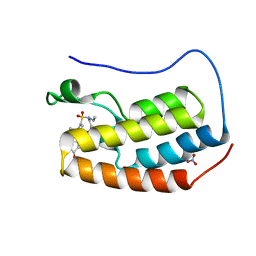 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 15c (N,N-diethyl-3',4'-dimethoxy-6-(1-methyl-5-oxopyrrolidin-3-yl)-[1,1'-biphenyl]-3-sulfonamide) | | Descriptor: | Bromodomain-containing protein 4, N,N-diethyl-3',4'-dimethoxy-6-[(3S)-1-methyl-5-oxopyrrolidin-3-yl][1,1'-biphenyl]-3-sulfonamide, NITRATE ION | | Authors: | Ilyichova, O.V, Scanlon, M.J, Thompson, P.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substituted 1-methyl-4-phenylpyrrolidin-2-ones - Fragment-based design of N-methylpyrrolidone-derived bromodomain inhibitors.
Eur.J.Med.Chem., 191, 2020
|
|
6VUC
 
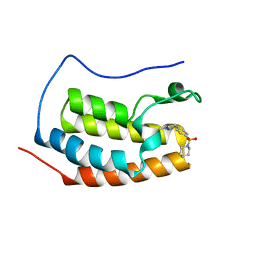 | |
3SL4
 
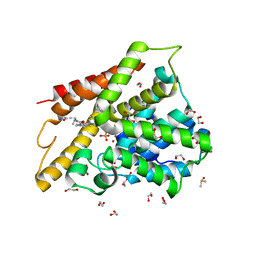 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL6
 
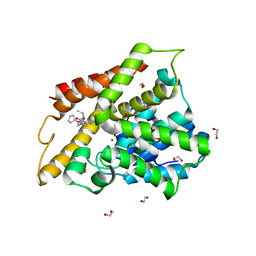 | | Crystal structure of the catalytic domain of PDE4D2 with compound 12c | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL8
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL3
 
 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL5
 
 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6PRT
 
 | |
6PS9
 
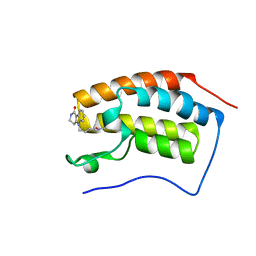 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 17 (5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one) | | Descriptor: | 5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one, Bromodomain-containing protein 4 | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Synthesis and elaboration of N-methylpyrrolidone as an acetamide fragment substitute in bromodomain inhibition.
Bioorg.Med.Chem., 27, 2019
|
|
6PSB
 
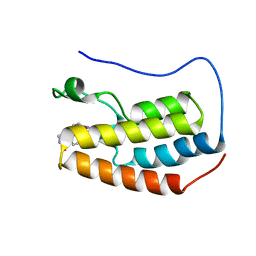 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 18 (5-{[(3R)-1-methyl-5-oxopyrrolidin-3-yl]methyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one) | | Descriptor: | 5-{[(3R)-1-methyl-5-oxopyrrolidin-3-yl]methyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one, Bromodomain-containing protein 4 | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Synthesis and elaboration of N-methylpyrrolidone as an acetamide fragment substitute in bromodomain inhibition.
Bioorg.Med.Chem., 27, 2019
|
|
4Y1C
 
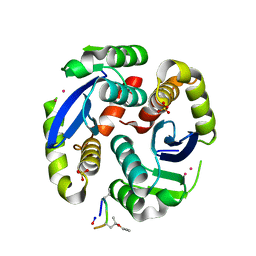 | |
4Y1D
 
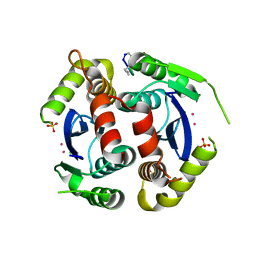 | |
4Z0E
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038TRN] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0F
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038(6CW)] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0D
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038Trp] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z09
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Thr2040Ala] peptide | | Descriptor: | Apical membrane antigen 1, GLYCEROL, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4OGB
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 2 | | Descriptor: | (2R)-8-(3,4-dimethoxyphenyl)-6-methyl-2-(tetrahydro-2H-pyran-4-yl)-2H-chromen-4-ol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feil, S.C, Parker, M.W. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | The PDE inhibition profile of LY294002 and tetrahydropyranyl analogues reveals a chromone motif for the development of PDE inhibitors
To be Published
|
|
4OVU
 
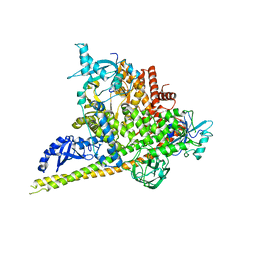 | | Crystal Structure of p110alpha in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
4OVV
 
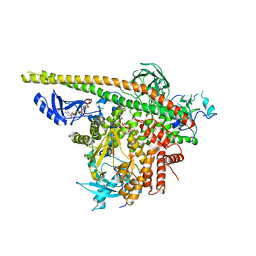 | | Crystal Structure of PI3Kalpha in complex with diC4-PIP2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
6DN7
 
 | |
6DN6
 
 | |
