1YPN
 
 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 1998-06-26 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
242D
 
 | | MAD PHASING STRATEGIES EXPLORED WITH A BROMINATED OLIGONUCLEOTIDE CRYSTAL AT 1.65 A RESOLUTION. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(CBR)P*G)-3') | | Authors: | Peterson, M.R, Harrop, S.J, McSweeney, S.M, Leonard, G.A, Thompson, A.W, Hunter, W.N, Helliwell, J.R. | | Deposit date: | 1996-06-20 | | Release date: | 1996-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MAD Phasing Strategies Explored with a Brominated Oligonucleotide Crystal at 1.65A Resolution.
J.Synchrotron Radiat., 3, 1996
|
|
19HC
 
 | | NINE-HAEM CYTOCHROME C FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ACETATE ION, PROTEIN (NINE-HAEM CYTOCHROME C), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Coelho, R, Pereira, I.A.C, Coelho, A.V, Thompson, A.W, Sieker, L, Gall, J.L, Carrondo, M.A. | | Deposit date: | 1998-12-01 | | Release date: | 1999-12-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family.
Structure Fold.Des., 7, 1999
|
|
3FOE
 
 | | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-8-chloro-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*AP*CP*CP*AP*AP*GP*GP*TP*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Laponogov, I, Sohi, M.K, Veselkov, D.A, Pan, X.-S, Sawhney, R, Thompson, A.W, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2008-12-30 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Nat.Struct.Mol.Biol., 16, 2009
|
|
3FOF
 
 | | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*AP*CP*CP*AP*AP*GP*GP*TP*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Laponogov, I, Sohi, M.K, Veselkov, D.A, Pan, X.-S, Sawhney, R, Thompson, A.W, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2008-12-30 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases
Nat.Struct.Mol.Biol., 16, 2009
|
|
4A4P
 
 | |
1GTK
 
 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1AH5
 
 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1ZEN
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Descriptor: | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE, ZINC ION | | Authors: | Cooper, S.J, Leonard, G.A, Hunter, W.N. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
Structure, 4, 1996
|
|
1JPO
 
 | | LOW TEMPERATURE ORTHORHOMBIC LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Bradbrook, G.M, Helliwell, J.R, Habash, J. | | Deposit date: | 1997-07-03 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Time-Resolved Biological and Perturbation Chemical Crystallography: Laue and Monochromatic Developments
Time-Resolved Electron and X-Ray Diffraction; 13-14 July 1995, San Diego, California (in: Proc.Spie-Int.Soc.Opt.Eng., V.2521), 1995
|
|
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILC
 
 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILA
 
 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Cesium | | Descriptor: | ACETATE ION, CESIUM ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IL4
 
 | | The pentameric ligand-gated ion channel GLIC in complex with Se-DDM | | Descriptor: | ACETATE ION, CHLORIDE ION, Proton-gated ion channel, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IL9
 
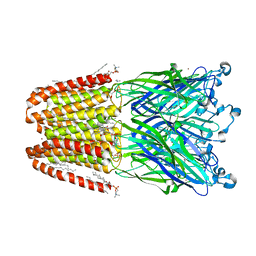 | | The pentameric ligand-gated ion channel GLIC A237F in complex with bromide | | Descriptor: | ACETATE ION, BROMIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILB
 
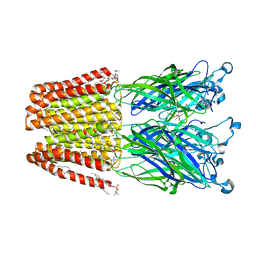 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Rubidium | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
1B57
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCOLOHYDROXAMIC ACID, PROTEIN (FRUCTOSE-BISPHOSPHATE ALDOLASE II), ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity.
J.Mol.Biol., 287, 1999
|
|
1B9J
 
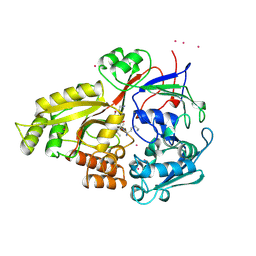 | |
1H21
 
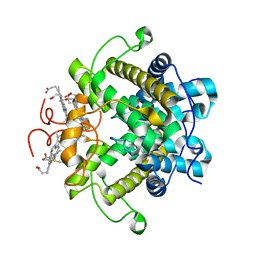 | | A novel iron centre in the split-Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | HEME C, SPLIT-SORET CYTOCHROME C | | Authors: | Abreu, I.A, Lourenco, A.I, Xavier, A.V, Legall, J, Coelho, A.V, Matias, P.M, Pinto, D.M, Carrondo, M.A, Teixeira, M, Saraiva, L.M. | | Deposit date: | 2002-07-30 | | Release date: | 2003-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Iron Centre in the Split-Soret Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774
J.Biol.Inorg.Chem., 8, 2003
|
|
1NF6
 
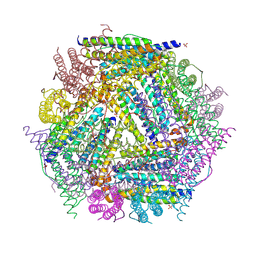 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF4
 
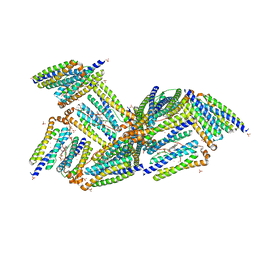 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
3K9F
 
 | | Detailed structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA (5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3'), ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
3KSB
 
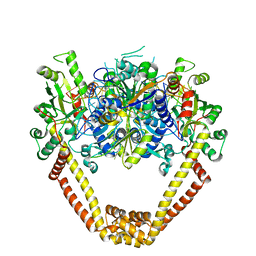 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (re-sealed form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*TP*GP*AP*CP*TP*AP*TP*GP*CP*AP*CP*GP*TP*AP*AP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*TP*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', DNA topoisomerase 4 subunit A, ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
3KSA
 
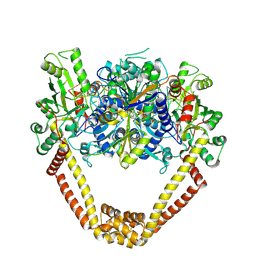 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (cleaved form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', 5'-D(P*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
