4RQO
 
 | |
1BXR
 
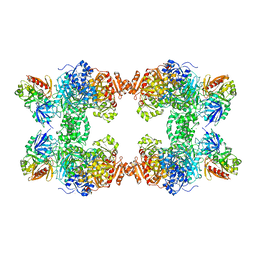 | | STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE COMPLEXED WITH THE ATP ANALOG AMPPNP | | Descriptor: | CARBAMOYL-PHOSPHATE SYNTHASE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Wesenberg, G, Raushel, F.M, Holden, H.M. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
Biochemistry, 38, 1999
|
|
1CE8
 
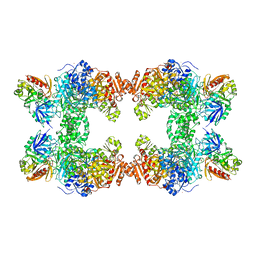 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIS COLI WITH COMPLEXED WITH THE ALLOSTERIC LIGAND IMP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, INOSINIC ACID, ... | | Authors: | Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-03-18 | | Release date: | 1999-07-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase.
J.Biol.Chem., 274, 1999
|
|
1CS0
 
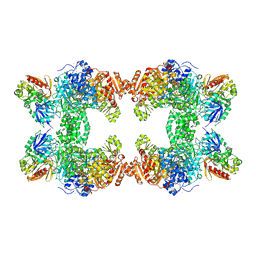 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1DV1
 
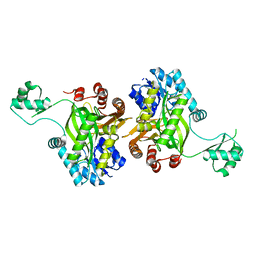 | | STRUCTURE OF BIOTIN CARBOXYLASE (APO) | | Descriptor: | BIOTIN CARBOXYLASE, PHOSPHATE ION | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
1SJC
 
 | | x-ray structure of o-succinylbenzoate synthase complexed with N-succinyl methionine | | Descriptor: | MAGNESIUM ION, N-SUCCINYL METHIONINE, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
1SJA
 
 | | X-ray structure of o-Succinylbenzoate Synthase complexed with N-acetylmethionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
1SJD
 
 | | x-ray structure of o-succinylbenzoate synthase complexed with n-succinyl phenylglycine | | Descriptor: | N-SUCCINYL PHENYLGLYCINE, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
1SJB
 
 | | X-ray structure of o-succinylbenzoate synthase complexed with o-succinylbenzoic acid | | Descriptor: | 2-SUCCINYLBENZOATE, MAGNESIUM ION, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
4EA8
 
 | | X-ray crystal structure of PerB from Caulobacter crescentus in complex with coenzyme A and GDP-N-acetylperosamine at 1 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-N-acetylperosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EAA
 
 | | X-ray crystal structure of the H141N mutant of perosamine N-acetyltransferase from Caulobacter crescentus in complex with CoA and GDP-perosamine | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EAB
 
 | | X-ray crystal structure of the H141A mutant of GDP-perosamine N-acetyl transferase from Caulobacter crescentus in complex with CoA and GDP-perosamine | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EA9
 
 | | X-ray structure of GDP-perosamine N-acetyltransferase in complex with transition state analog at 0.9 Angstrom resolution | | Descriptor: | CHLORIDE ION, GDP-N-acetylperosamine-coenzyme A, Perosamine N-acetyltransferase | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EA7
 
 | | X-ray crystal structure of PerB from Caulobacter crescentus in complex with CoA and GDP-perosamine at 1.0 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
1UDA
 
 | |
2A2C
 
 | |
1UDC
 
 | |
1UDB
 
 | |
2A2D
 
 | |
2AJ4
 
 | | Crystal structure of Saccharomyces cerevisiae Galactokinase in complex with galactose and Mg:AMPPNP | | Descriptor: | CHLORIDE ION, Galactokinase, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Sellick, C.A, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer
J.Biol.Chem., 280, 2005
|
|
3K5H
 
 | | Crystal structure of carboxyaminoimidazole ribonucleotide synthase from asperigillus clavatus complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosyl-aminoimidazole carboxylase | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
1WUU
 
 | | crystal structure of human galactokinase complexed with MgAMPPNP and galactose | | Descriptor: | Galactokinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Thoden, J.B, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Structure of Human Galactokinase: IMPLICATIONS FOR TYPE II GALACTOSEMIA
J.Biol.Chem., 280, 2005
|
|
3K5I
 
 | | Crystal structure of N5-carboxyaminoimidazole synthase from aspergillus clavatus in complex with ADP and 5-aminoimadazole ribonucleotide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
2E3D
 
 | |
3MQG
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with acetyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETYL COENZYME *A, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
