4AG5
 
 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WJT
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLK
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with reaction product | | Descriptor: | N-[(1R,2S,3R,4R,5R)-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-4-yl]ethanamide, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
4WLI
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis | | Descriptor: | 1,2-ETHANEDIOL, YuiC | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-07 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
2MHQ
 
 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2MHP
 
 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
4AG6
 
 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | SULFATE ION, TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A8A
 
 | | Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-28 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (14.2 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A9G
 
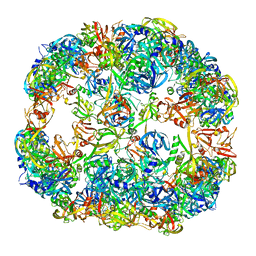 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8B
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8C
 
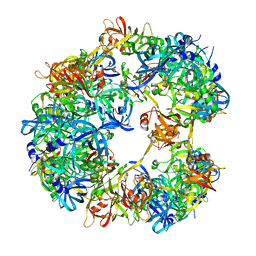 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
7PDJ
 
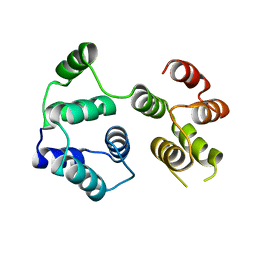 | | R12E vFLIP mutant | | Descriptor: | FLICE inhibitory protein | | Authors: | Barrett, T.E. | | Deposit date: | 2021-08-05 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into the activation of the IKK kinase complex by the Kaposi's sarcoma herpes virus oncoprotein vFLIP.
J.Biol.Chem., 298, 2022
|
|
5O0U
 
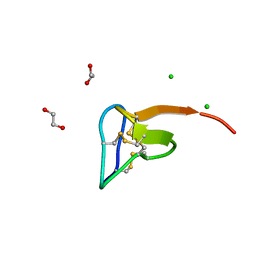 | | Crystal structure of tarantula venom peptide Protoxin-II | | Descriptor: | 1,2-ETHANEDIOL, Beta/omega-theraphotoxin-Tp2a, CHLORIDE ION | | Authors: | Tabor, A, McCarthy, S, Reyes, F.E. | | Deposit date: | 2017-05-17 | | Release date: | 2017-09-13 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | The Role of Disulfide Bond Replacements in Analogues of the Tarantula Toxin ProTx-II and Their Effects on Inhibition of the Voltage-Gated Sodium Ion Channel Nav1.7.
J.Am.Chem.Soc., 139, 2017
|
|
4PYW
 
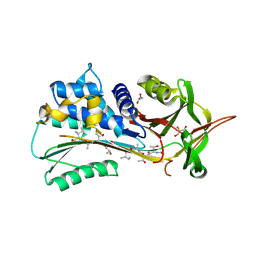 | | 1.92 angstrom crystal structure of A1AT:TTAI ternary complex | | Descriptor: | ACE-THR-THR-ALA-ILE-NH2, Alpha-1-antitrypsin, GLYCEROL | | Authors: | Nyon, M.P, Day, J, Gooptu, B. | | Deposit date: | 2014-03-28 | | Release date: | 2015-06-10 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An integrative approach combining ion mobility mass spectrometry, X-ray crystallography, and nuclear magnetic resonance spectroscopy to study the conformational dynamics of alpha 1 -antitrypsin upon ligand binding.
Protein Sci., 24, 2015
|
|
8BDK
 
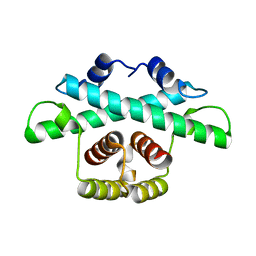 | |
5AN3
 
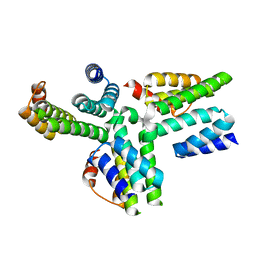 | | Structure of an Sgt1-Skp1 Complex | | Descriptor: | SGT1, SUPPRESSOR OF KINETOCHORE PROTEIN 1 | | Authors: | Willhoft, O, Vaughan, C.K. | | Deposit date: | 2015-09-03 | | Release date: | 2017-02-08 | | Last modified: | 2017-02-15 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The crystal structure of the Sgt1-Skp1 complex: the link between Hsp90 and both SCF E3 ubiquitin ligases and kinetochores.
Sci Rep, 7, 2017
|
|
2YMV
 
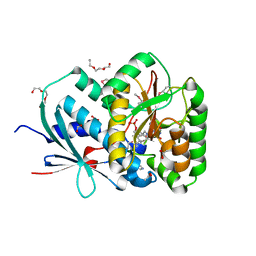 | | Structure of Reduced M Smegmatis 5246, a homologue of M.Tuberculosis Acg | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, ACG NITROREDUCTASE, ... | | Authors: | Chauviac, F.-X, Bommer, M, Dobbek, H, Keep, N.H. | | Deposit date: | 2012-10-10 | | Release date: | 2012-11-21 | | Last modified: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Reduced Msacg, a Putative Nitroreductase from Mycobacterium Smegmatis and a Close Homologue of Mycobacterium Tuberculosis Acg.
J.Biol.Chem., 287, 2012
|
|
4AAY
 
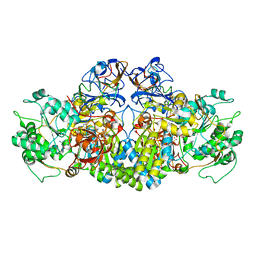 | | Crystal Structure of the arsenite oxidase protein complex from Rhizobium species strain NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AROA, AROB, ... | | Authors: | Oke, M, Santini, J.M, Naismith, J.H. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Respiratory Arsenite Oxidase: Structure and the Role of Residues Surrounding the Rieske Cluster.
Plos One, 8, 2013
|
|
8BA7
 
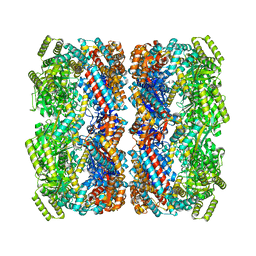 | |
8BA8
 
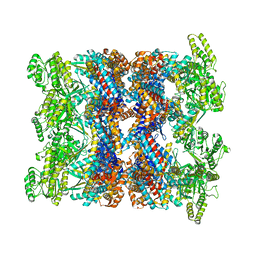 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BA9
 
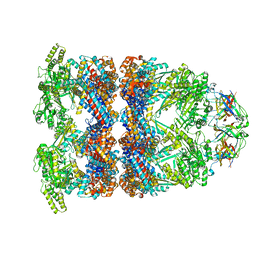 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
6GYZ
 
 | |
6GYW
 
 | |
6GYY
 
 | |
6GYX
 
 | |
