1DWN
 
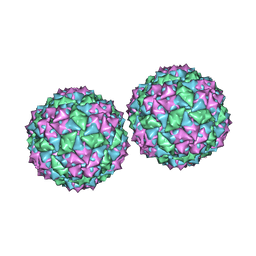 | | Structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7 A resolution | | Descriptor: | PHAGE COAT PROTEIN | | Authors: | Tars, K, Fridborg, K, Bundule, M, Liljas, L. | | Deposit date: | 1999-12-09 | | Release date: | 2000-02-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure Determination of Phage Pp7 from Pseudomonas Aeruginosa: From Poor Data to a Good Map
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1GAV
 
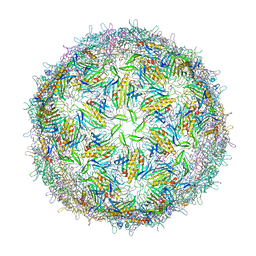 | | BACTERIOPHAGE GA PROTEIN CAPSID | | Descriptor: | BACTERIOPHAGE GA PROTEIN CAPSID | | Authors: | Tars, K, Bundule, M, Liljas, L. | | Deposit date: | 1997-01-28 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses.
J.Mol.Biol., 271, 1997
|
|
7Q6P
 
 | |
1NG0
 
 | |
2VCT
 
 | |
2VCV
 
 | |
2WJU
 
 | |
2YKL
 
 | | Structure of human anti-nicotine Fab fragment in complex with nicotine-11-yl-methyl-(4-ethylamino-4-oxo)-butanoate | | Descriptor: | FAB FRAGMENT, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Tars, K, Kotelovica, S, Lipowsky, G, Bauer, M, Beerli, R, Bachmann, M, Maurer, P. | | Deposit date: | 2011-05-27 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Binding Modes of Free and Carrier-Protein-Coupled Nicotine in a Human Monoclonal Antibody.
J.Mol.Biol., 415, 2012
|
|
2YK1
 
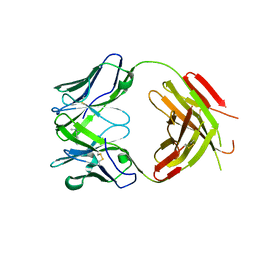 | | Structure of human anti-nicotine Fab fragment in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, FAB FRAGMENT, HEAVY CHAIN, ... | | Authors: | Tars, K, Kotelovica, S, Lipowsky, G, Bauer, M, Beerli, R, Bachmann, M, Maurer, P. | | Deposit date: | 2011-05-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Different Binding Modes of Free and Carrier-Protein-Coupled Nicotine in a Human Monoclonal Antibody.
J.Mol.Biol., 415, 2012
|
|
4BHI
 
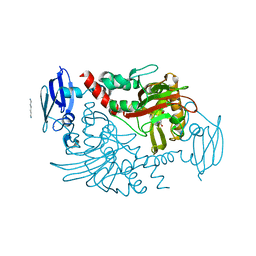 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1,1,1,2-Tetramethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1,1,1,2-TETRAMETHYLHYDRAZIN-1-IUM-2-YL)PROPANOATE, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BHF
 
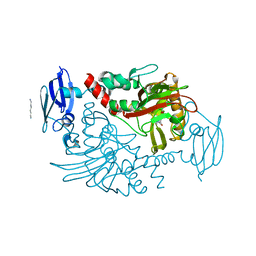 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 4-(Trimethylammonio)pentanoate | | Descriptor: | 4-(Trimethylammonio)pentanoic acid, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BGM
 
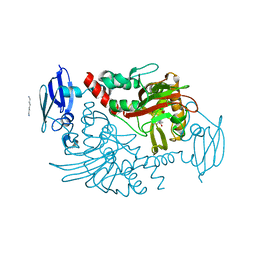 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-Carboxy-N-(2-fluoroethyl)-N,N-dimethylpropan-1- aminium chloride | | Descriptor: | 3-Carboxy-N-(2-fluoroethyl)-N,N-dimethylpropan-1-aminium, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BHG
 
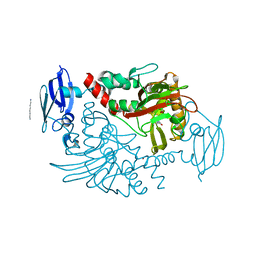 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1-Ethyl-1,1-dimethylhydrazin-1-ium-2-yl)propanoic acid, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three Dimensional Structure of Human Gamma-Butyrobetaine Hydroxylase in Complex with 3-(1-Ethyl-1,1-Dimethylhydrazin-1-Ium-2-Yl)Propanoate
To be Published
|
|
4BGK
 
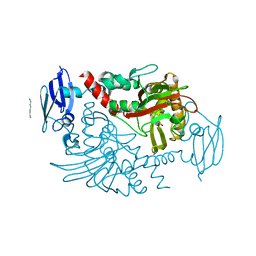 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with (3-(Trimethylammonio)propyl)phosphinate | | Descriptor: | GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, N-OXALYLGLYCINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BG1
 
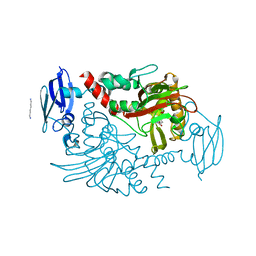 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 1-(3-Carboxypropyl)-1-methylpyrrolidin-1-ium chloride | | Descriptor: | 1-(3-carboxypropyl)-1-methylpyrrolidin-1-ium, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BCW
 
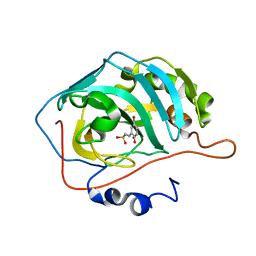 | | Carbonic anhydrase IX mimic in complex with (E)-2-(5-bromo-2- hydroxyphenyl)ethenesulfonic acid | | Descriptor: | (E)-2-(5-BROMO-2-HYDROXYPHENYL)ETHENESULFONIC ACID, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfocoumarins (1,2-Benzoxathiine-2,2-Dioxides): A Class of Potent and Isoform-Selective Inhibitors of Tumor-Associated Carbonic Anhydrases.
J.Med.Chem., 56, 2013
|
|
4C5W
 
 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 4-(Ethyldimethylammonio)butanoate | | Descriptor: | GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, N-OXALYLGLYCINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
8OG0
 
 | |
2C3Q
 
 | | Human glutathione-S-transferase T1-1 W234R mutant, complex with S- hexylglutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2C3T
 
 | | Human glutathione-S-transferase T1-1, W234R mutant, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1 | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-12 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2C3N
 
 | | Human glutathione-S-transferase T1-1, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
7OOM
 
 | |
3N6W
 
 | | Crystal structure of human gamma-butyrobetaine hydroxylase | | Descriptor: | Gamma-butyrobetaine dioxygenase, SULFATE ION, ZINC ION | | Authors: | Rumnieks, J, Zeltins, A, Leonchiks, A, Kazaks, A, Kotelovica, S, Tars, K. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human gamma-butyrobetaine hydroxylase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
4CKU
 
 | | Three dimensional structure of plasmepsin II in complex with hydroxyethylamine-based inhibitor | | Descriptor: | 5-[1,1-bis(oxidanylidene)-1,2-thiazinan-2-yl]-N3-[(2S,3R)-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-butan-2-yl]-N1,N1-dipropyl-benzene-1,3-dicarboxamide, PLASMEPSIN-2 | | Authors: | Tars, K, Leitans, J, Jaudzems, K. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasmepsin Inhibitory Activity and Structure-Guided Optimization of a Potent Hydroxyethylamine-Based Antimalarial Hit.
Acs Med.Chem.Lett., 5, 2014
|
|
2J9H
 
 | | Crystal structure of human glutathione-S-transferase P1-1 cys-free mutant in complex with S-hexylglutathione at 2.4 A resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE P, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Hegazy, U.M, Hellman, U, Mannervik, B. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modulating Catalytic Activity by Unnatural Amino Acid Residues in a Gsh-Binding Loop of Gst P1-1.
J.Mol.Biol., 376, 2008
|
|
