6UQR
 
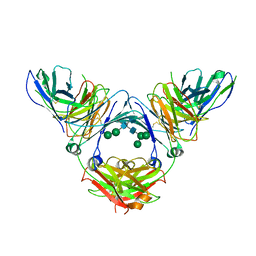 | | Complex of IgE and Ligelizumab | | Descriptor: | IgE, Ligelizumab, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tarchevskaya, S.S, Kleinboelting, S, Jardetzky, T.S. | | Deposit date: | 2019-10-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6502912 Å) | | Cite: | The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab.
Nat Commun, 11, 2020
|
|
1F6A
 
 | | Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Wurzburg, B.A, Tarchevskaya, S.S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc (epsilon) RI (alpha).
Nature, 406, 2000
|
|
5HYS
 
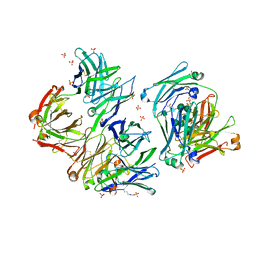 | | Structure of IgE complexed with omalizumab | | Descriptor: | Epididymis luminal protein 214, Ig epsilon chain C region, SULFATE ION, ... | | Authors: | Pennington, L.F, Tarchevskaya, S.S, Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange.
Nat Commun, 7, 2016
|
|
4GRG
 
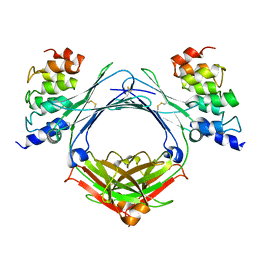 | |
4GT7
 
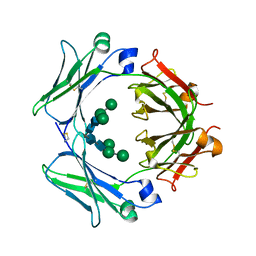 | | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, non-receptor binding conformation | | Descriptor: | Ig epsilon chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wurzburg, B.A, Kim, B.K, Jardetzky, T.S. | | Deposit date: | 2012-08-28 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An engineered disulfide bond reversibly traps the IgE-Fc3-4 in a closed, nonreceptor binding conformation.
J.Biol.Chem., 287, 2012
|
|
2H2R
 
 | | Crystal structure of the human CD23 Lectin domain, apo form | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII)(Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
2H2T
 
 | | CD23 Lectin domain, Calcium 2+-bound | | Descriptor: | CALCIUM ION, Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
