1IWQ
 
 | | Crystal Structure of MARCKS calmodulin binding domain peptide complexed with Ca2+/Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, MARCKS | | Authors: | Yamauchi, E, Nakatsu, T, Matsubara, M, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-31 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca(2+)-calmodulin
NAT.STRUCT.BIOL., 10, 2003
|
|
1WSV
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1WSR
 
 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1L7Z
 
 | | Crystal structure of Ca2+/Calmodulin complexed with myristoylated CAP-23/NAP-22 peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CAP-23/NAP-22, ... | | Authors: | Matsubara, M, Nakatsu, T, Yamauchi, E, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-18 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin
EMBO J., 23, 2004
|
|
3W82
 
 | | Human alpha-L-iduronidase in complex with iduronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W81
 
 | | Human alpha-l-iduronidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1X2H
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
5ZZ4
 
 | | Crystal structure of bruton's tyrosine kinase in complex with inhibitor 2e | | Descriptor: | N-[3-(4-amino-6-{[4-(morpholine-4-carbonyl)phenyl]amino}-1,3,5-triazin-2-yl)-2-methylphenyl]-4-tert-butylbenzamide, Tyrosine-protein kinase BTK | | Authors: | Kawahata, W, Asami, T, Irie, T, Kiyoi, T, Taniguchi, H, Asamitsu, Y, Inoue, T, Miyake, T, Sawa, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and Synthesis of Novel Amino-triazine Analogues as Selective Bruton's Tyrosine Kinase Inhibitors for Treatment of Rheumatoid Arthritis.
J. Med. Chem., 61, 2018
|
|
3WD4
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor and quinoline compound | | Descriptor: | (E)-N-(prop-2-en-1-yloxy)-1-(quinolin-4-yl)methanimine, Chitinase B, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD2
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD1
 
 | | Serratia marcescens Chitinase B complexed with syn-triazole inhibitor | | Descriptor: | Chitinase B, GLYCEROL, N,N-dibenzyl-N~5~-[N-(methylcarbamoyl)carbamimidoyl]-N~2~-{[5-({[(E)-(quinolin-4-ylmethylidene)amino]oxy}methyl)-1H-1,2,3-triazol-1-yl]acetyl}-L-ornithinamide, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD3
 
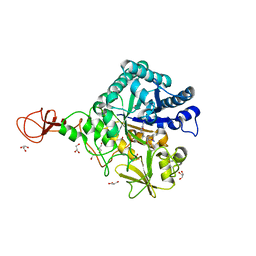 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, GLYCEROL, SULFATE ION, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD0
 
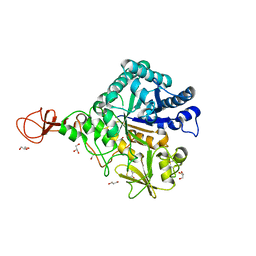 | | Serratia marcescens Chitinase B, tetragonal form | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7DPJ
 
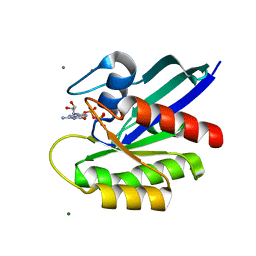 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | Authors: | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
1A2X
 
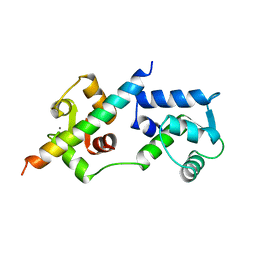 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4Z2L
 
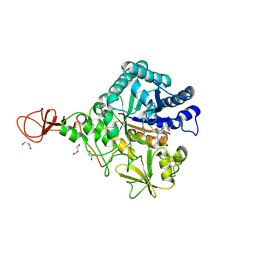 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 33 | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-14-ethyl-7,10,12,13-tetrahydroxy-3,5,7,9,11,13-hexamethyl-2-oxo-6-(prop-2-yn-1-yloxy)oxacyclotetradecan-4-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2G
 
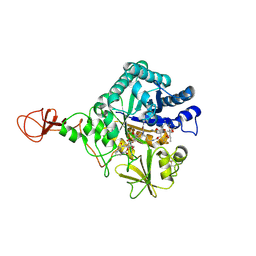 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 26 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {4-[N'-(methylcarbamoyl)carbamimidamido]butyl}carbamate, Chitinase B | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2J
 
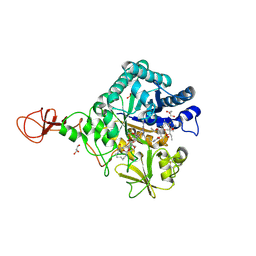 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 31 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {5-[N'-(methylcarbamoyl)carbamimidamido]pentyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2H
 
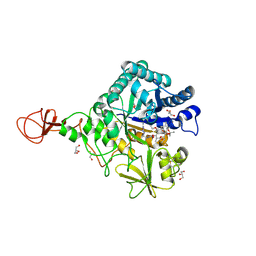 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 29 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {2-[N'-(methylcarbamoyl)carbamimidamido]ethyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2K
 
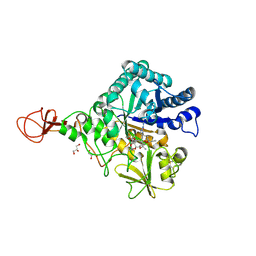 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2I
 
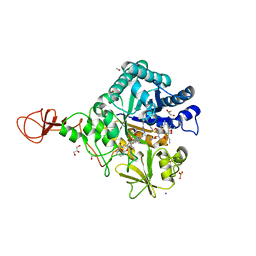 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 30 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
3A7A
 
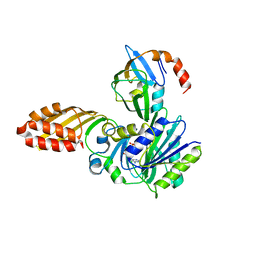 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
