3FF5
 
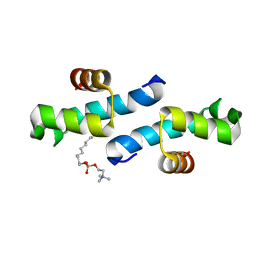 | | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix-protein-import receptor, Pex14p | | Descriptor: | Peroxisomal biogenesis factor 14, decyl 2-trimethylazaniumylethyl phosphate | | Authors: | Su, J.-R, Takeda, K, Tamura, S, Fujiki, Y, Miki, K. | | Deposit date: | 2008-12-01 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2A4G
 
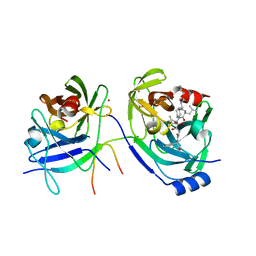 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2ZW2
 
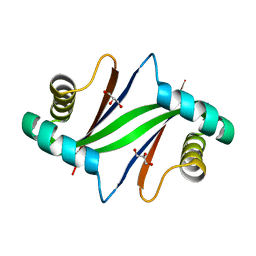 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
3VIU
 
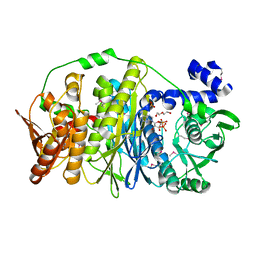 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KJO
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the low affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
2Z04
 
 | | Crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit from Aquifex aeolicus | | Descriptor: | Phosphoribosylaminoimidazole carboxylase ATPase subunit, SULFATE ION | | Authors: | Okada, K, Tamura, S, Baba, S, Kanagawa, M, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit from Aquifex aeolicus
To be Published
|
|
4YQM
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-27 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-N-[4-chloro-3-(dimethylsulfamoyl)phenyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YQV
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C4-10 | | Descriptor: | 2-(ethylsulfanyl)-N-methyl-N-[(1-phenyl-1H-pyrazol-4-yl)methyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YQU
 
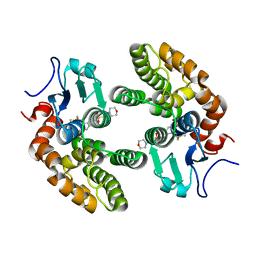 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-31 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-{5-(azepan-1-ylsulfonyl)-2-[(ethylsulfanyl)methoxy]phenyl}acetamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
3AX6
 
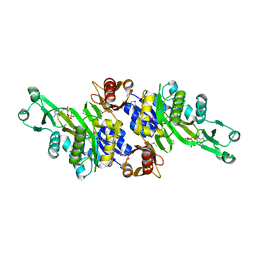 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Miyazawa, R, Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
6KIQ
 
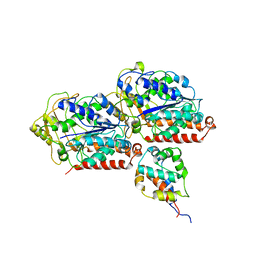 | | Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT | | Descriptor: | Alpha tubulin, Dynein heavy chain, cytoplasmic, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KIO
 
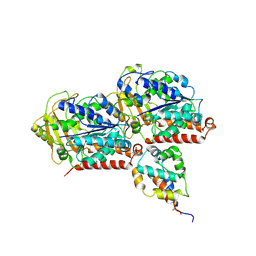 | | Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT | | Descriptor: | Dynein heavy chain, cytoplasmic, Tubulin alpha-1A chain, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
2A4R
 
 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6CJG
 
 | | Human dihydroorotate dehydrogenase bound to napthyridine inhibitor 46 | | Descriptor: | 2-([1,1'-biphenyl]-4-yl)-3-methyl-1,7-naphthyridine-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6CJF
 
 | | Human dihydroorotate dehydrogenase bound to 4-quinoline carboxylic acid inhibitor 43 | | Descriptor: | 2-[4-(2-chloro-6-methylpyridin-3-yl)phenyl]-6-fluoro-3-methylquinoline-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
