3J04
 
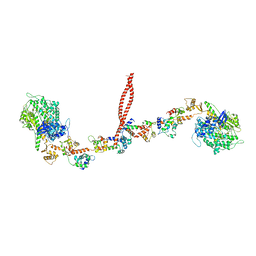 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
4C3G
 
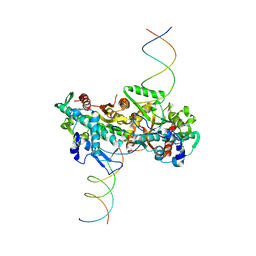 | | cryo-EM structure of activated and oligomeric restriction endonuclease SgrAI | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*TP*GP*AP*AP*GP*AP*CP*CP *CP*AP*CP*GP*CP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*GP*CP*GP*TP*GP*GP*GP*TP*CP*TP*TP *CP*AP*CP*AP)-3', SGRAIR RESTRICTION ENZYME | | Authors: | Lyumkis, D, Talley, H, Stewart, A, Shah, S, Park, C.K, Tama, F, Potter, C.S, Carragher, B, Horton, N.C. | | Deposit date: | 2013-08-23 | | Release date: | 2013-09-11 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Allosteric Regulation of DNA Cleavage and Sequence-Specificity Through Run-on Oligomerization.
Structure, 21, 2013
|
|
2AKI
 
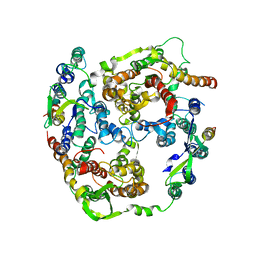 | | Normal mode-based flexible fitted coordinates of a translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
2AKH
 
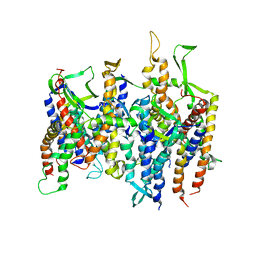 | | Normal mode-based flexible fitted coordinates of a non-translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K.M, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
6K8Q
 
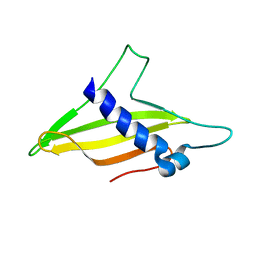 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6OF7
 
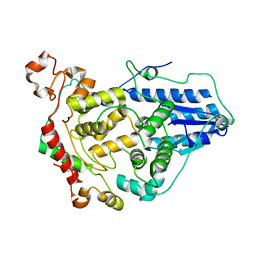 | | Crystal structure of the CRY1-PER2 complex | | Descriptor: | Cryptochrome-1, Period circadian protein homolog 2 | | Authors: | Michael, A.K, Fribourgh, J.L, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Elife, 9, 2020
|
|
4V4W
 
 | |
4V4V
 
 | |
7WSV
 
 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7V8Y
 
 | | Crystal structure of mouse CRY2 in complex with SHP1703 compound | | Descriptor: | 1-[(2R)-3-[3,6-bis(fluoranyl)carbazol-9-yl]-2-oxidanyl-propyl]imidazolidin-2-one, Cryptochrome-2 | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8Z
 
 | | Crystal structure of mouse CRY2 in complex with SHP656 compound | | Descriptor: | 1-[(2R)-3-[3,6-bis(fluoranyl)carbazol-9-yl]-2-oxidanyl-propyl]imidazolidin-2-one, Cryptochrome-2 | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6A1C
 
 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
6DTL
 
 | | Mitogen-activated protein kinase 6 | | Descriptor: | Mitogen-activated protein kinase 6 | | Authors: | Ruble, J. | | Deposit date: | 2018-06-17 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS.
Nat.Plants, 5, 2019
|
|
7D1C
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with compound TH303 | | Descriptor: | Cryptochrome-1, N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]-4-(phenylcarbonyl)benzamide | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2020-09-14 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Photopharmacological Manipulation of Mammalian CRY1 for Regulation of the Circadian Clock.
J.Am.Chem.Soc., 143, 2021
|
|
7D19
 
 | |
7D0N
 
 | | Crystal structure of mouse CRY2 apo form | | Descriptor: | Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DLI
 
 | | Crystal structure of mouse CRY1 in complex with KL001 compound | | Descriptor: | 1,2-ETHANEDIOL, Cryptochrome-1, N-[(2R)-3-carbazol-9-yl-2-oxidanyl-propyl]-N-(furan-2-ylmethyl)methanesulfonamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-11-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D0M
 
 | | Crystal structure of mouse CRY1 with bound cryoprotectant | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EJ9
 
 | | Alternative crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6K7F
 
 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K7D
 
 | |
6K7E
 
 | |
